Saif-ur-Rehman, Muhammad; Ali, Omair; Klaes, Christian; Iossifidis, Ioannis In: Neurocomputing, S. 131370, 2025, ISSN: 0925-2312. Abstract | Links | BibTeX | Schlagwörter: BCI, Brain computer interface, Deep learning, Self organizing, Self-supervised machine learning, Spike Sorting Ali, Omair; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian In: Computers in Biology and Medicine, S. 107649, 2023, ISSN: 0010-4825. Abstract | Links | BibTeX | Schlagwörter: BCI, Brain computer interface, Deep learning, EEG decoding, EMG decoding, Machine Learning2025
@article{saif-ur-rehmanAdaptiveSpikeDeepclassifierSelforganizing2025,
title = {Adaptive SpikeDeep-classifier: Self-organizing and Self-Supervised Machine Learning Algorithm for Online Spike Sorting},
author = {Muhammad Saif-ur-Rehman and Omair Ali and Christian Klaes and Ioannis Iossifidis},
editor = {Elsevier},
url = {https://www.sciencedirect.com/science/article/pii/S0925231225020429},
doi = {10.1016/j.neucom.2025.131370},
issn = {0925-2312},
year = {2025},
date = {2025-09-04},
urldate = {2025-09-04},
journal = {Neurocomputing},
pages = {131370},
abstract = {Objective. Invasive brain-computer interface (BCI) research is progressing towards the realization of the motor skills rehabilitation of severely disabled patients in the real world. The size of invasive micro-electrode arrays and the selection of an efficient online spike sorting algorithm (performing spike sorting at run time) are two key factors that play pivotal roles in the successful decoding of the user’s intentions. The process of spike sorting includes the selection of channels that record the spike activity (SA) and determines the SA of different sources (neurons), on selected channels individually. The neural data recorded with dense micro-electrode arrays is time-varying and often contaminated with non-stationary noise. Unfortunately, current state-of-the-art spike sorting algorithms are incapable of handling the massively increasing amount of time-varying data resulting from the dense microelectrode arrays, which makes the spike sorting one of the fragile components of the online BCI decoding framework. Approach. This study proposed an adaptive and self-organized algorithm for online spike sorting, named as Adaptive SpikeDeep-Classifier (Ada-SpikeDeepClassifier). Our algorithm uses SpikeDeeptector for the channel selection, an adaptive background activity rejector (Ada-BAR) for discarding the background events, and an adaptive spike deep-classifier (Ada-SpikeDeepClassifier) for classifying the SA of different neural units. The process of spike sorting is accomplished by concatenating SpikeDeeptector, Ada-BAR and Ada-SpikeDeepclassifier. Results. The proposed algorithm is evaluated on two different categories of data: a human data-set recorded in our lab, and a publicly available simulated data-set to avoid subjective biases and labeling errors. The proposed Ada-SpikeDeepClassifier outperformed our previously published SpikeDeep-Classifier and eight conventional spike sorting algorithms and produce comparable results to state of the art deep learning based algorithms. Significance. To the best of our knowledge, the proposed algorithm is the first spike sorting algorithm that autonomously adapts to the shift in the distribution of noise and SA data and perform spike sorting without human interventions in various kinds of experimental settings. In addition, the proposed algorithm builds upon artificial neural networks, which makes it an ideal candidate for being embedded on neuromorphic chips that are also suitable for wearable invasive BCI.},
keywords = {BCI, Brain computer interface, Deep learning, Self organizing, Self-supervised machine learning, Spike Sorting},
pubstate = {published},
tppubtype = {article}
}
2023
@article{aliConTraNetHybridNetwork2023,
title = {ConTraNet: A Hybrid Network for Improving the Classification of EEG and EMG Signals with Limited Training Data},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {https://www.sciencedirect.com/science/article/pii/S0010482523011149},
doi = {10.1016/j.compbiomed.2023.107649},
issn = {0010-4825},
year = {2023},
date = {2023-11-02},
urldate = {2023-11-02},
journal = {Computers in Biology and Medicine},
pages = {107649},
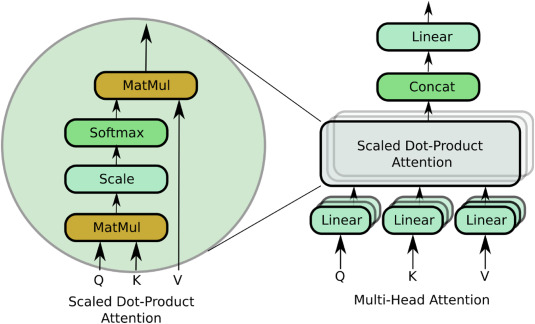
abstract = {Objective Bio-Signals such as electroencephalography (EEG) and electromyography (EMG) are widely used for the rehabilitation of physically disabled people and for the characterization of cognitive impairments. Successful decoding of these bio-signals is however non-trivial because of the time-varying and non-stationary characteristics. Furthermore, existence of short- and long-range dependencies in these time-series signal makes the decoding even more challenging. State-of-the-art studies proposed Convolutional Neural Networks (CNNs) based architectures for the classification of these bio-signals, which are proven useful to learn spatial representations. However, CNNs because of the fixed size convolutional kernels and shared weights pay only uniform attention and are also suboptimal in learning short-long term dependencies, simultaneously, which could be pivotal in decoding EEG and EMG signals. Therefore, it is important to address these limitations of CNNs. To learn short- and long-range dependencies simultaneously and to pay more attention to more relevant part of the input signal, Transformer neural network-based architectures can play a significant role. Nonetheless, it requires a large corpus of training data. However, EEG and EMG decoding studies produce limited amount of the data. Therefore, using standalone transformers neural networks produce ordinary results. In this study, we ask a question whether we can fix the limitations of CNN and transformer neural networks and provide a robust and generalized model that can simultaneously learn spatial patterns, long-short term dependencies, pay variable amount of attention to time-varying non-stationary input signal with limited training data. Approach In this work, we introduce a novel single hybrid model called ConTraNet, which is based on CNN and Transformer architectures that contains the strengths of both CNN and Transformer neural networks. ConTraNet uses a CNN block to introduce inductive bias in the model and learn local dependencies, whereas the Transformer block uses the self-attention mechanism to learn the short- and long-range or global dependencies in the signal and learn to pay different attention to different parts of the signals. Main results We evaluated and compared the ConTraNet with state-of-the-art methods on four publicly available datasets (BCI Competition IV dataset 2b, Physionet MI-EEG dataset, Mendeley sEMG dataset, Mendeley sEMG V1 dataset) which belong to EEG-HMI and EMG-HMI paradigms. ConTraNet outperformed its counterparts in all the different category tasks (2-class, 3-class, 4-class, 7-class, and 10-class decoding tasks). Significance With limited training data ConTraNet significantly improves classification performance on four publicly available datasets for 2, 3, 4, 7, and 10-classes compared to its counterparts.},
keywords = {BCI, Brain computer interface, Deep learning, EEG decoding, EMG decoding, Machine Learning},
pubstate = {published},
tppubtype = {article}
}