Lehrgebiet: Theoretische Informatik und künstliche Intelligenz
Büro: 01.214
Labor: 04.105
Telefon: +49 208 88254-806
E-Mail:
🛜 http://lab.iossifidis.net

Ioannis Iossifidis studierte Physik (Schwerpunkt: theoretische Teilchenphysik) an der Universität Dortmund und promovierte 2006 an der Fakultät für Physik und Astronomie der Ruhr-Universität Bochum.
Am Institut für Neuroinformatik leitete Prof. Dr. Iossifidis die Arbeitsgruppe Autonome Robotik und nahm mit seiner Forschungsgruppe erfolgreich an zahlreichen, vom BmBF und der EU, geförderten Forschungsprojekten aus dem Bereich der künstlichen Intelligenz teil. Seit dem 1. Oktober 2010 arbeitet er an der HRW am Institut Informatik und hält den Lehrstuhl für Theoretische Informatik – Künstliche Intelligenz.
Prof. Dr. Ioannis Iossifidis entwickelt seit über 20 Jahren biologisch inspirierte anthropomorphe, autonome Robotersysteme, die zugleich Teil und Ergebnis seiner Forschung im Bereich der rechnergestützten Neurowissenschaften sind. In diesem Rahmen entwickelte er Modelle zur Informationsverarbeitung im menschlichen Gehirn und wendete diese auf technische Systeme an.
Ausgewiesene Schwerpunkte seiner wissenschaftlichen Arbeit der letzten Jahre sind die Modellierung menschlicher Armbewegungen, der Entwurf von sogenannten «Simulierten Realitäten» zur Simulation und Evaluation der Interaktionen zwischen Mensch, Maschine und Umwelt sowie die Entwicklung von kortikalen exoprothetischen Komponenten. Entwicklung der Theorie und Anwendung von Algorithmen des maschinellen Lernens auf Basis tiefer neuronaler Architekturen bilden das Querschnittsthema seiner Forschung.
Ioannis Iossifidis’ Forschung wurde u.a. mit Fördermitteln im Rahmen großer Förderprojekte des BmBF (NEUROS, MORPHA, LOKI, DESIRE, Bernstein Fokus: Neuronale Grundlagen des Lernens etc.), der DFG («Motor‐parietal cortical neuroprosthesis with somatosensory feedback for restoring hand and arm functions in tetraplegic patients») und der EU (Neural Dynamics – EU (STREP), EUCogII, EUCogIII ) honoriert und gehört zu den Gewinnern der Leitmarktwettbewerbe Gesundheit.NRW und IKT.NRW 2019.
ARBEITS- UND FORSCHUNGSSCHWERPUNKTE
- Computational Neuroscience
- Brain Computer Interfaces
- Entwicklung kortikaler exoprothetischer Komponenten
- Theorie neuronaler Netze
- Modellierung menschlicher Armbewegungen
- Simulierte Realität
WISSENSCHAFTLICHE EINRICHTUNGEN
- Labor mit Verlinkung
- ???
- ???
LEHRVERANSTALTUNGEN
- ???
- ???
- ???
PROJEKTE
- Projekt mit Verlinkung
- ???
- ???
WISSENSCHAFTLICHE MITARBEITER*INNEN

Felix Grün
Büro: 02.216 (Campus Bottrop)

Marie Schmidt
Büro: 02.216 (Campus Bottrop)

Aline Xavier Fidencio
Gastwissenschaftlerin

Muhammad Ayaz Hussain
Doktorand

Tim Sziburis
Doktorand

Farhad Rahmat
studentische Hilfskraft
GOOGLE SCHOLAR PROFIL

Artikel
Fidêncio, Aline Xavier; Grün, Felix; Klaes, Christian; Iossifidis, Ioannis
Hybrid Brain-Computer Interface Using Error-Related Potential and Reinforcement Learning Artikel
In: Frontiers in Human Neuroscience, Bd. 19, 2025, ISSN: 1662-5161.
Abstract | Links | BibTeX | Schlagwörter: adaptive brain-computer interface, BCI, EEG, error-related potentials (ErrPs), Machine Learning, motor imagery (MI), reinforcement learning (RL)
@article{xavierfidencioHybridBraincomputerInterface2025,
title = {Hybrid Brain-Computer Interface Using Error-Related Potential and Reinforcement Learning},
author = {Aline Xavier Fidêncio and Felix Grün and Christian Klaes and Ioannis Iossifidis},
editor = {Frontiers},
url = {https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2025.1569411/full},
doi = {10.3389/fnhum.2025.1569411},
issn = {1662-5161},
year = {2025},
date = {2025-06-04},
urldate = {2025-06-04},
journal = {Frontiers in Human Neuroscience},
volume = {19},
publisher = {Frontiers},
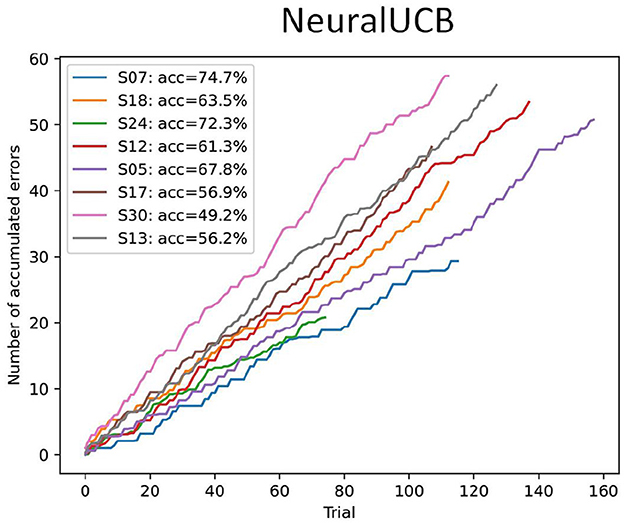
abstract = {Brain-computer interfaces (BCIs) offer alternative communication methods for individuals with motor disabilities, aiming to improve their quality of life through external device control. However, non-invasive BCIs using electroencephalography (EEG) often suffer from performance limitations due to non-stationarities arising from changes in mental state or device characteristics. Addressing these challenges motivates the development of adaptive systems capable of real-time adjustment. This study investigates a novel approach for creating an adaptive, error-related potential (ErrP)-based BCI using reinforcement learning (RL) to dynamically adapt to EEG signal variations. The framework was validated through experiments on a publicly available motor imagery dataset and a novel fast-paced protocol designed to enhance user engagement. Results showed that RL agents effectively learned control policies from user interactions, maintaining robust performance across datasets. However, findings from the game-based protocol revealed that fast-paced motor imagery tasks were ineffective for most participants, highlighting critical challenges in real-time BCI task design. Overall, the results demonstrate the potential of RL for enhancing BCI adaptability while identifying practical constraints in task complexity and user responsiveness.},
keywords = {adaptive brain-computer interface, BCI, EEG, error-related potentials (ErrPs), Machine Learning, motor imagery (MI), reinforcement learning (RL)},
pubstate = {published},
tppubtype = {article}
}
Fidêncio, Aline Xavier; Grün, Felix; Klaes, Christian; Iossifidis, Ioannis
Error-Related Potential Driven Reinforcement Learning for Adaptive Brain-Computer Interfaces Artikel
In: Arxiv, 2025.
Abstract | Links | BibTeX | Schlagwörter: BCI, Computer Science - Human-Computer Interaction, Computer Science - Machine Learning, EEG, Quantitative Biology - Neurons and Cognition, Reinforcement learning
@article{fidencioErrorrelatedPotentialDriven2025,
title = {Error-Related Potential Driven Reinforcement Learning for Adaptive Brain-Computer Interfaces},
author = {Aline Xavier Fidêncio and Felix Grün and Christian Klaes and Ioannis Iossifidis},
url = {http://arxiv.org/abs/2502.18594},
doi = {10.48550/arXiv.2502.18594},
year = {2025},
date = {2025-02-25},
urldate = {2025-02-25},
journal = {Arxiv},
abstract = {Brain-computer interfaces (BCIs) provide alternative communication methods for individuals with motor disabilities by allowing control and interaction with external devices. Non-invasive BCIs, especially those using electroencephalography (EEG), are practical and safe for various applications. However, their performance is often hindered by EEG non-stationarities, caused by changing mental states or device characteristics like electrode impedance. This challenge has spurred research into adaptive BCIs that can handle such variations. In recent years, interest has grown in using error-related potentials (ErrPs) to enhance BCI performance. ErrPs, neural responses to errors, can be detected non-invasively and have been integrated into different BCI paradigms to improve performance through error correction or adaptation. This research introduces a novel adaptive ErrP-based BCI approach using reinforcement learning (RL). We demonstrate the feasibility of an RL-driven adaptive framework incorporating ErrPs and motor imagery. Utilizing two RL agents, the framework adapts dynamically to EEG non-stationarities. Validation was conducted using a publicly available motor imagery dataset and a fast-paced game designed to boost user engagement. Results show the framework's promise, with RL agents learning control policies from user interactions and achieving robust performance across datasets. However, a critical insight from the game-based protocol revealed that motor imagery in a high-speed interaction paradigm was largely ineffective for participants, highlighting task design limitations in real-time BCI applications. These findings underscore the potential of RL for adaptive BCIs while pointing out practical constraints related to task complexity and user responsiveness.},
keywords = {BCI, Computer Science - Human-Computer Interaction, Computer Science - Machine Learning, EEG, Quantitative Biology - Neurons and Cognition, Reinforcement learning},
pubstate = {published},
tppubtype = {article}
}
Lehmler, Stephan Johann; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis
In: Neurocomputing, S. 128473, 2024, ISSN: 0925-2312.
Abstract | Links | BibTeX | Schlagwörter: Artificial neural networks, Generalization, Machine Learning, Memorization, Poisson process, Stochastic modeling
@article{lehmlerUnderstandingActivationPatterns2024,
title = {Understanding Activation Patterns in Artificial Neural Networks by Exploring Stochastic Processes: Discriminating Generalization from Memorization},
author = {Stephan Johann Lehmler and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis},
editor = {Elsevier},
url = {https://www.sciencedirect.com/science/article/pii/S092523122401244X},
doi = {10.1016/j.neucom.2024.128473},
issn = {0925-2312},
year = {2024},
date = {2024-09-19},
urldate = {2024-09-19},
journal = {Neurocomputing},
pages = {128473},
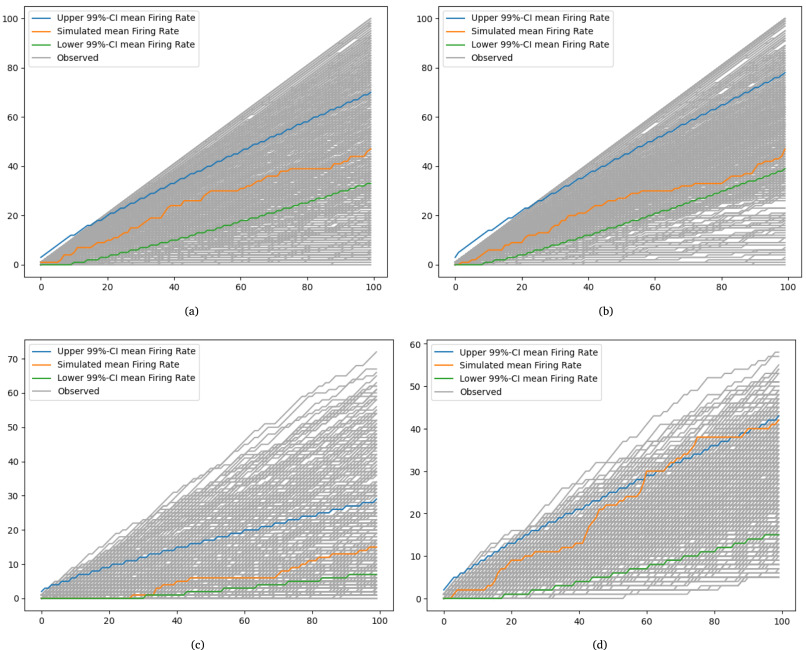
abstract = {To gain a deeper understanding of the behavior and learning dynamics of artificial neural networks, mathematical abstractions and models are valuable. They provide a simplified perspective and facilitate systematic investigations. In this paper, we propose to analyze dynamics of artificial neural activation using stochastic processes, which have not been utilized for this purpose thus far. Our approach involves modeling the activation patterns of nodes in artificial neural networks as stochastic processes. By focusing on the activation frequency, we can leverage techniques used in neuroscience to study neural spike trains. Specifically, we extract the activity of individual artificial neurons during a classification task and model their activation frequency. The underlying process model is an arrival process following a Poisson distribution.We examine the theoretical fit of the observed data generated by various artificial neural networks in image recognition tasks to the proposed model’s key assumptions. Through the stochastic process model, we derive measures describing activation patterns of each network. We analyze randomly initialized, generalizing, and memorizing networks, allowing us to identify consistent differences in learning methods across multiple architectures and training sets. We calculate features describing the distribution of Activation Rate and Fano Factor, which prove to be stable indicators of memorization during learning. These calculated features offer valuable insights into network behavior. The proposed model demonstrates promising results in describing activation patterns and could serve as a general framework for future investigations. It has potential applications in theoretical simulation studies as well as practical areas such as pruning or transfer learning.},
keywords = {Artificial neural networks, Generalization, Machine Learning, Memorization, Poisson process, Stochastic modeling},
pubstate = {published},
tppubtype = {article}
}
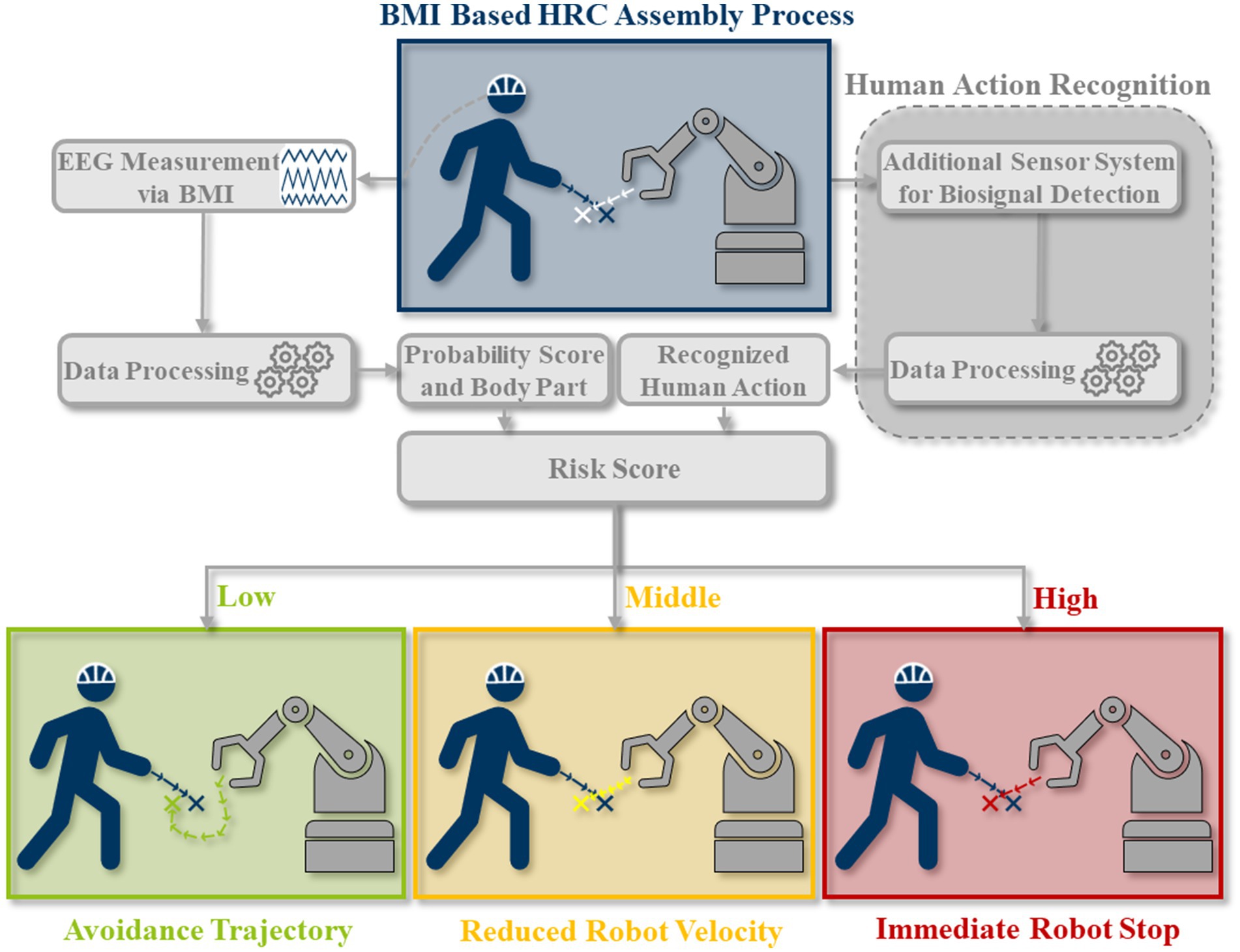
Pilacinski, Artur; Christ, Lukas; Boshoff, Marius; Iossifidis, Ioannis; Adler, Patrick; Miro, Michael; Kuhlenkötter, Bernd; Klaes, Christian
In: Frontiers in Neurorobotics, Bd. 18, 2024, ISSN: 1662-5218.
Links | BibTeX | Schlagwörter: brain-machine interfaces, EEG, Human action recognition, human-robot collaboration, Sensor Fusion
@article{pilacinskiHumanCollaborativeLoop2024,
title = {Human in the Collaborative Loop: A Strategy for Integrating Human Activity Recognition and Non-Invasive Brain-Machine Interfaces to Control Collaborative Robots},
author = {Artur Pilacinski and Lukas Christ and Marius Boshoff and Ioannis Iossifidis and Patrick Adler and Michael Miro and Bernd Kuhlenkötter and Christian Klaes},
url = {https://www.frontiersin.org/journals/neurorobotics/articles/10.3389/fnbot.2024.1383089/full},
doi = {10.3389/fnbot.2024.1383089},
issn = {1662-5218},
year = {2024},
date = {2024-09-18},
urldate = {2024-09-18},
journal = {Frontiers in Neurorobotics},
volume = {18},
publisher = {Frontiers},
keywords = {brain-machine interfaces, EEG, Human action recognition, human-robot collaboration, Sensor Fusion},
pubstate = {published},
tppubtype = {article}
}
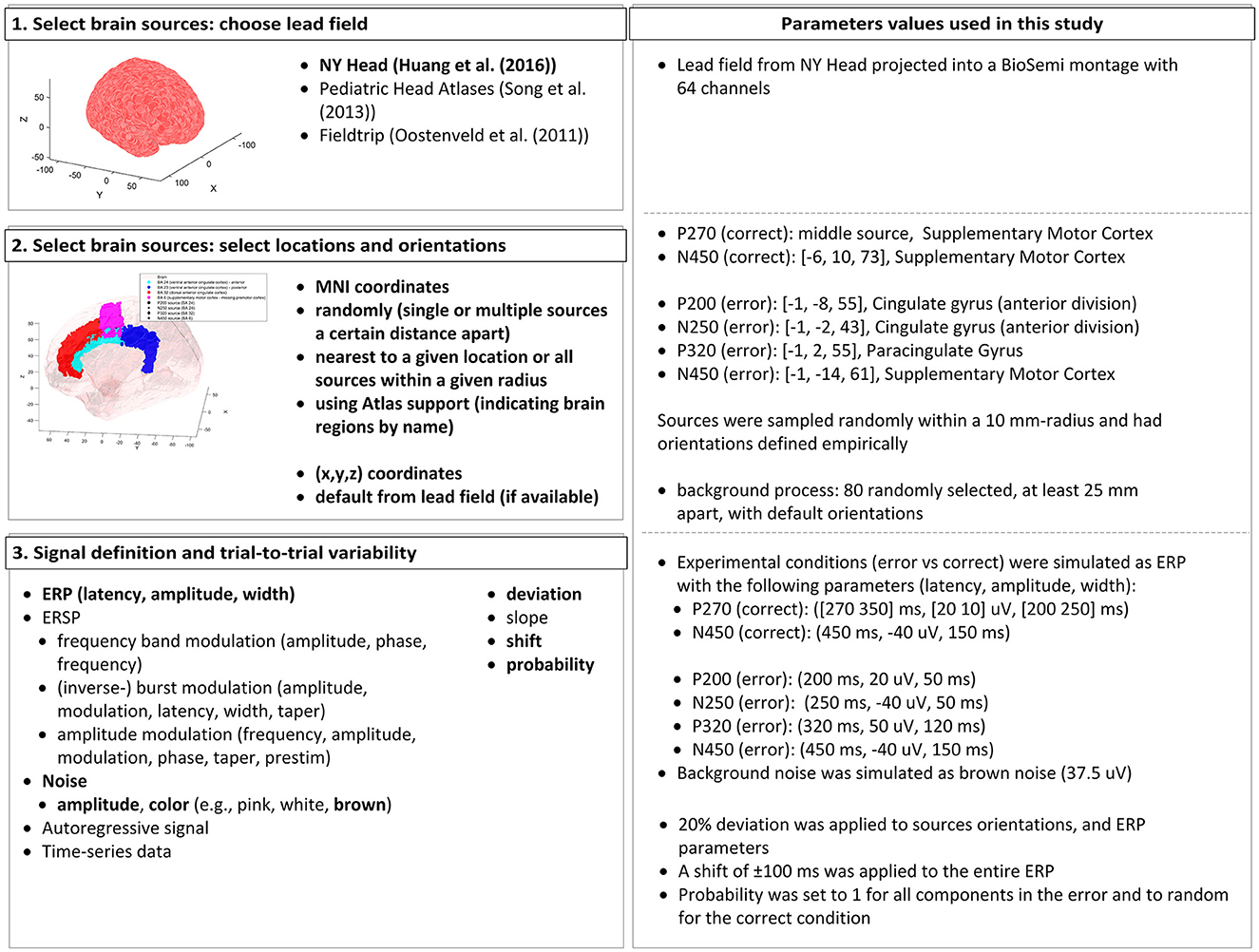
Fidêncio, Aline Xavier; Klaes, Christian; Iossifidis, Ioannis
A Generic Error-Related Potential Classifier Based on Simulated Subjects Artikel
In: Frontiers in Human Neuroscience, Bd. 18, S. 1390714, 2024, ISSN: 1662-5161.
Abstract | Links | BibTeX | Schlagwörter: adaptive brain-machine (computer) interface, BCI, EEG, Error-related potential (ErrP), ErrP classifier, Generic decoder, Machine Learning, SEREEGA, Simulation
@article{xavierfidencioGenericErrorrelatedPotential2024,
title = {A Generic Error-Related Potential Classifier Based on Simulated Subjects},
author = {Aline Xavier Fidêncio and Christian Klaes and Ioannis Iossifidis},
editor = {Frontiers Media SA},
url = {https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2024.1390714/full},
doi = {10.3389/fnhum.2024.1390714},
issn = {1662-5161},
year = {2024},
date = {2024-07-19},
urldate = {2024-07-19},
journal = {Frontiers in Human Neuroscience},
volume = {18},
pages = {1390714},
publisher = {Frontiers},
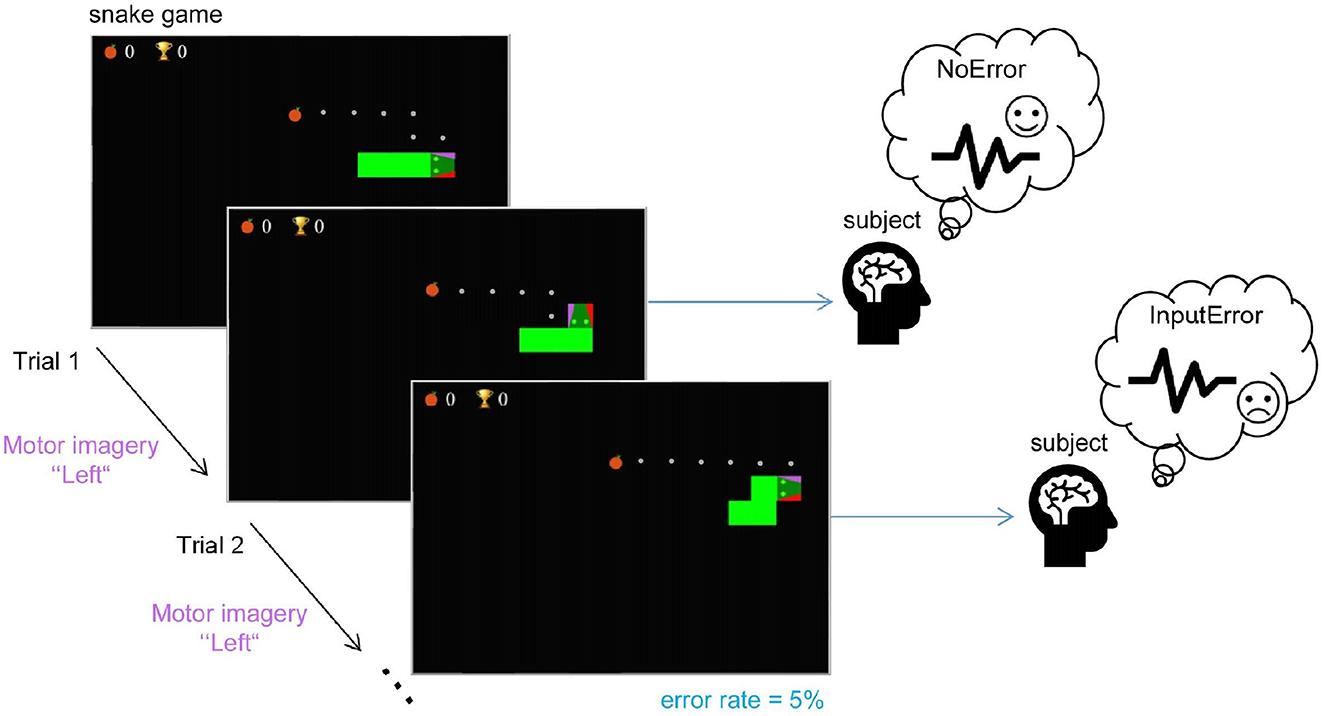
abstract = {$<$p$>$Error-related potentials (ErrPs) are brain signals known to be generated as a reaction to erroneous events. Several works have shown that not only self-made errors but also mistakes generated by external agents can elicit such event-related potentials. The possibility of reliably measuring ErrPs through non-invasive techniques has increased the interest in the brain-computer interface (BCI) community in using such signals to improve performance, for example, by performing error correction. Extensive calibration sessions are typically necessary to gather sufficient trials for training subject-specific ErrP classifiers. This procedure is not only time-consuming but also boresome for participants. In this paper, we explore the effectiveness of ErrPs in closed-loop systems, emphasizing their dependency on precise single-trial classification. To guarantee the presence of an ErrPs signal in the data we employ and to ensure that the parameters defining ErrPs are systematically varied, we utilize the open-source toolbox SEREEGA for data simulation. We generated training instances and evaluated the performance of the generic classifier on both simulated and real-world datasets, proposing a promising alternative to conventional calibration techniques. Results show that a generic support vector machine classifier reaches balanced accuracies of 72.9%, 62.7%, 71.0%, and 70.8% on each validation dataset. While performing similarly to a leave-one-subject-out approach for error class detection, the proposed classifier shows promising generalization across different datasets and subjects without further adaptation. Moreover, by utilizing SEREEGA, we can systematically adjust parameters to accommodate the variability in the ErrP, facilitating the systematic validation of closed-loop setups. Furthermore, our objective is to develop a universal ErrP classifier that captures the signal's variability, enabling it to determine the presence or absence of an ErrP in real EEG data.$<$/p$>$},
keywords = {adaptive brain-machine (computer) interface, BCI, EEG, Error-related potential (ErrP), ErrP classifier, Generic decoder, Machine Learning, SEREEGA, Simulation},
pubstate = {published},
tppubtype = {article}
}
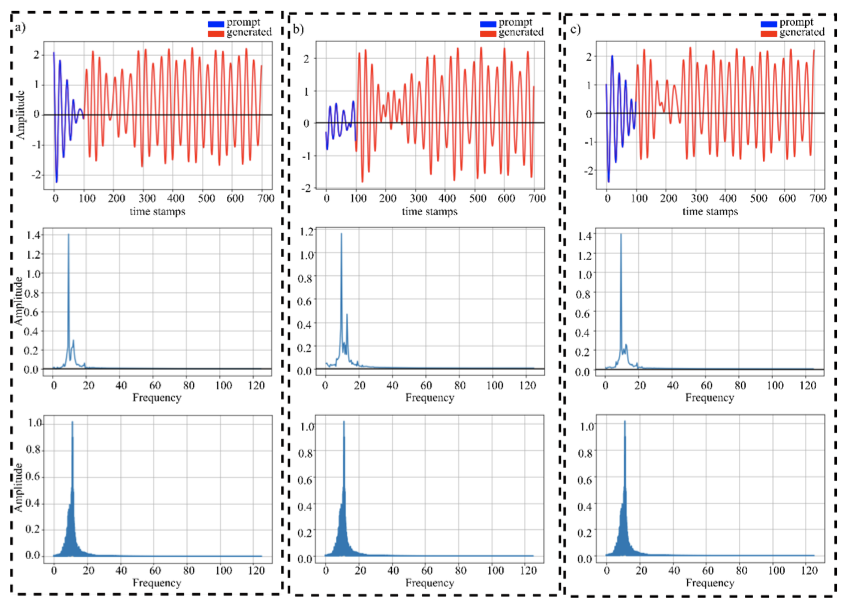
Ali, Omair; Saif-ur-Rehman, Muhammad; Metzler, Marita; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian
GET: A Generative EEG Transformer for Continuous Context-Based Neural Signals Artikel
In: arXiv:2406.03115 [q-bio], 2024.
Abstract | Links | BibTeX | Schlagwörter: BCI, EEG, Machine Learning, Quantitative Biology - Neurons and Cognition
@article{aliGETGenerativeEEG2024,
title = {GET: A Generative EEG Transformer for Continuous Context-Based Neural Signals},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Marita Metzler and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {http://arxiv.org/abs/2406.03115},
doi = {10.48550/arXiv.2406.03115},
year = {2024},
date = {2024-06-09},
urldate = {2024-06-09},
journal = {arXiv:2406.03115 [q-bio]},
abstract = {Generating continuous electroencephalography (EEG) signals through advanced artificial neural networks presents a novel opportunity to enhance brain-computer interface (BCI) technology. This capability has the potential to significantly enhance applications ranging from simulating dynamic brain activity and data augmentation to improving real-time epilepsy detection and BCI inference. By harnessing generative transformer neural networks, specifically designed for EEG signal generation, we can revolutionize the interpretation and interaction with neural data. Generative AI has demonstrated significant success across various domains, from natural language processing (NLP) and computer vision to content creation in visual arts and music. It distinguishes itself by using large-scale datasets to construct context windows during pre-training, a technique that has proven particularly effective in NLP, where models are fine-tuned for specific downstream tasks after extensive foundational training. However, the application of generative AI in the field of BCIs, particularly through the development of continuous, context-rich neural signal generators, has been limited. To address this, we introduce the Generative EEG Transformer (GET), a model leveraging transformer architecture tailored for EEG data. The GET model is pre-trained on diverse EEG datasets, including motor imagery and alpha wave datasets, enabling it to produce high-fidelity neural signals that maintain contextual integrity. Our empirical findings indicate that GET not only faithfully reproduces the frequency spectrum of the training data and input prompts but also robustly generates continuous neural signals. By adopting the successful training strategies of the NLP domain for BCIs, the GET sets a new standard for the development and application of neural signal generation technologies.},
keywords = {BCI, EEG, Machine Learning, Quantitative Biology - Neurons and Cognition},
pubstate = {published},
tppubtype = {article}
}
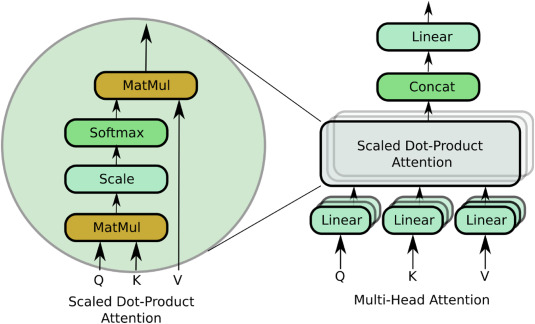
Ali, Omair; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian
In: Computers in Biology and Medicine, S. 107649, 2023, ISSN: 0010-4825.
Abstract | Links | BibTeX | Schlagwörter: BCI, Brain computer interface, Deep learning, EEG decoding, EMG decoding, Machine Learning
@article{aliConTraNetHybridNetwork2023,
title = {ConTraNet: A Hybrid Network for Improving the Classification of EEG and EMG Signals with Limited Training Data},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {https://www.sciencedirect.com/science/article/pii/S0010482523011149},
doi = {10.1016/j.compbiomed.2023.107649},
issn = {0010-4825},
year = {2023},
date = {2023-11-02},
urldate = {2023-11-02},
journal = {Computers in Biology and Medicine},
pages = {107649},
abstract = {Objective Bio-Signals such as electroencephalography (EEG) and electromyography (EMG) are widely used for the rehabilitation of physically disabled people and for the characterization of cognitive impairments. Successful decoding of these bio-signals is however non-trivial because of the time-varying and non-stationary characteristics. Furthermore, existence of short- and long-range dependencies in these time-series signal makes the decoding even more challenging. State-of-the-art studies proposed Convolutional Neural Networks (CNNs) based architectures for the classification of these bio-signals, which are proven useful to learn spatial representations. However, CNNs because of the fixed size convolutional kernels and shared weights pay only uniform attention and are also suboptimal in learning short-long term dependencies, simultaneously, which could be pivotal in decoding EEG and EMG signals. Therefore, it is important to address these limitations of CNNs. To learn short- and long-range dependencies simultaneously and to pay more attention to more relevant part of the input signal, Transformer neural network-based architectures can play a significant role. Nonetheless, it requires a large corpus of training data. However, EEG and EMG decoding studies produce limited amount of the data. Therefore, using standalone transformers neural networks produce ordinary results. In this study, we ask a question whether we can fix the limitations of CNN and transformer neural networks and provide a robust and generalized model that can simultaneously learn spatial patterns, long-short term dependencies, pay variable amount of attention to time-varying non-stationary input signal with limited training data. Approach In this work, we introduce a novel single hybrid model called ConTraNet, which is based on CNN and Transformer architectures that contains the strengths of both CNN and Transformer neural networks. ConTraNet uses a CNN block to introduce inductive bias in the model and learn local dependencies, whereas the Transformer block uses the self-attention mechanism to learn the short- and long-range or global dependencies in the signal and learn to pay different attention to different parts of the signals. Main results We evaluated and compared the ConTraNet with state-of-the-art methods on four publicly available datasets (BCI Competition IV dataset 2b, Physionet MI-EEG dataset, Mendeley sEMG dataset, Mendeley sEMG V1 dataset) which belong to EEG-HMI and EMG-HMI paradigms. ConTraNet outperformed its counterparts in all the different category tasks (2-class, 3-class, 4-class, 7-class, and 10-class decoding tasks). Significance With limited training data ConTraNet significantly improves classification performance on four publicly available datasets for 2, 3, 4, 7, and 10-classes compared to its counterparts.},
keywords = {BCI, Brain computer interface, Deep learning, EEG decoding, EMG decoding, Machine Learning},
pubstate = {published},
tppubtype = {article}
}

Hussain, Muhammad Ayaz; Iossifidis, Ioannis
In: arXiv:2309.04698 [cs.RO], 2023.
Abstract | Links | BibTeX | Schlagwörter: Autonomous robotics, BCI, Computer Science - Artificial Intelligence, Computer Science - Information Theory, Computer Science - Machine Learning, Exoskeleton
@article{ayazhussainAdvancementsUpperBody2023,
title = {Advancements in Upper Body Exoskeleton: Implementing Active Gravity Compensation with a Feedforward Controller},
author = {Muhammad Ayaz Hussain and Ioannis Iossifidis},
url = {https://doi.org/10.48550/arXiv.2309.04698},
doi = {10.48550/arXiv.2309.04698},
year = {2023},
date = {2023-09-09},
urldate = {2023-09-09},
journal = {arXiv:2309.04698 [cs.RO]},
abstract = {In this study, we present a feedforward control system designed for active gravity compensation on an upper body exoskeleton. The system utilizes only positional data from internal motor sensors to calculate torque, employing analytical control equations based on Newton-Euler Inverse Dynamics. Compared to feedback control systems, the feedforward approach offers several advantages. It eliminates the need for external torque sensors, resulting in reduced hardware complexity and weight. Moreover, the feedforward control exhibits a more proactive response, leading to enhanced performance. The exoskeleton used in the experiments is lightweight and comprises 4 Degrees of Freedom, closely mimicking human upper body kinematics and three-dimensional range of motion. We conducted tests on both hardware and simulations of the exoskeleton, demonstrating stable performance. The system maintained its position over an extended period, exhibiting minimal friction and avoiding undesired slewing.},
keywords = {Autonomous robotics, BCI, Computer Science - Artificial Intelligence, Computer Science - Information Theory, Computer Science - Machine Learning, Exoskeleton},
pubstate = {published},
tppubtype = {article}
}
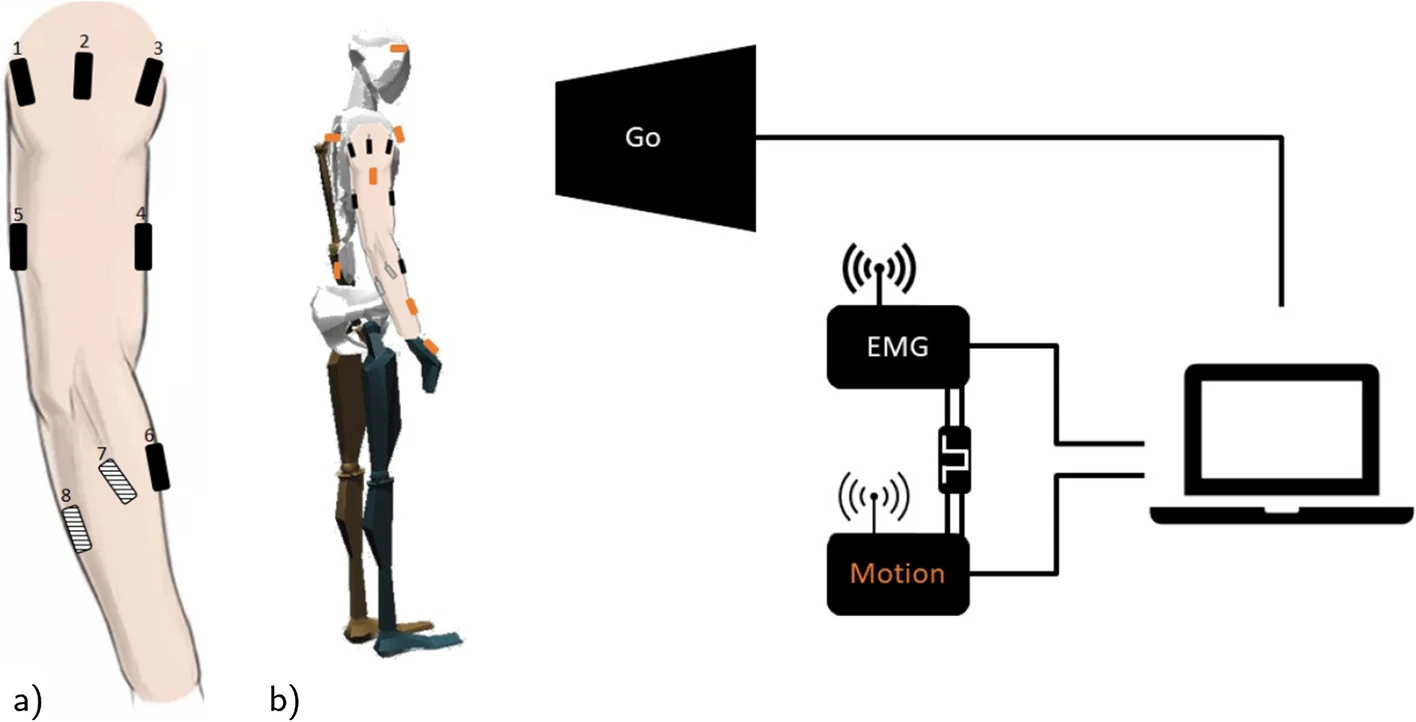
Schmidt, Marie D.; Glasmachers, Tobias; Iossifidis, Ioannis
The Concepts of Muscle Activity Generation Driven by Upper Limb Kinematics Artikel
In: BioMedical Engineering OnLine, Bd. 22, Nr. 1, S. 63, 2023, ISSN: 1475-925X.
Abstract | Links | BibTeX | Schlagwörter: Artificial generated signal, BCI, Electromyography (EMG), Generative model, Inertial measurement unit (IMU), Machine Learning, Motion parameters, Muscle activity, Neural networks, transfer learning, Voluntary movement
@article{schmidtConceptsMuscleActivity2023,
title = {The Concepts of Muscle Activity Generation Driven by Upper Limb Kinematics},
author = {Marie D. Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
url = {https://doi.org/10.1186/s12938-023-01116-9},
doi = {10.1186/s12938-023-01116-9},
issn = {1475-925X},
year = {2023},
date = {2023-06-24},
urldate = {2023-06-24},
journal = {BioMedical Engineering OnLine},
volume = {22},
number = {1},
pages = {63},
abstract = {The underlying motivation of this work is to demonstrate that artificial muscle activity of known and unknown motion can be generated based on motion parameters, such as angular position, acceleration, and velocity of each joint (or the end-effector instead), which are similarly represented in our brains. This model is motivated by the known motion planning process in the central nervous system. That process incorporates the current body state from sensory systems and previous experiences, which might be represented as pre-learned inverse dynamics that generate associated muscle activity.},
keywords = {Artificial generated signal, BCI, Electromyography (EMG), Generative model, Inertial measurement unit (IMU), Machine Learning, Motion parameters, Muscle activity, Neural networks, transfer learning, Voluntary movement},
pubstate = {published},
tppubtype = {article}
}
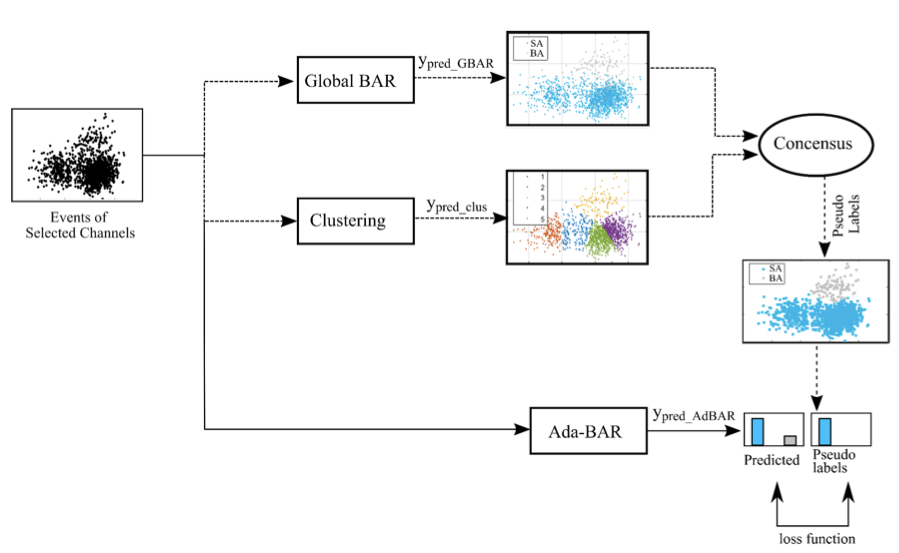
Saif-ur-Rehman, Muhammad; Ali, Omair; Klaes, Christian; Iossifidis, Ioannis
In: arXiv:2304.01355 [cs, math, q-bio], 2023.
Links | BibTeX | Schlagwörter: BCI, Machine Learning, Spike Sorting
@article{saifurrehman2023adaptive,
title = {Adaptive SpikeDeep-Classifier: Self-organizing and self-supervised machine learning algorithm for online spike sorting},
author = {Muhammad Saif-ur-Rehman and Omair Ali and Christian Klaes and Ioannis Iossifidis},
doi = {10.48550/arXiv.2304.01355},
year = {2023},
date = {2023-05-02},
urldate = {2023-05-02},
journal = {arXiv:2304.01355 [cs, math, q-bio]},
keywords = {BCI, Machine Learning, Spike Sorting},
pubstate = {published},
tppubtype = {article}
}