Schmidt, Marie D.; Glasmachers, Tobias; Iossifidis, Ioannis Insights into Motor Control: Predict Muscle Activity from Upper Limb Kinematics with LSTM Networks Artikel In: Nature Scientific Reports, 2026, ISSN: 2045-2322. Abstract | Links | BibTeX | Schlagwörter: BCI, Computational biology and bioinformatics, Motor control, Neuroscience Schmidt, Marie Dominique; Iossifidis, Ioannis Predicting Upper Limb Muscle Activity from Kinematics Using LSTM Networks Proceedings Article In: Bernstein Conference 2025, BCCN Bernstein Network Computational Neuroscience & Neurotechnology , 2025. Abstract | Links | BibTeX | Schlagwörter: BCI Fidencio, Aline Xavier; Iossifidis, Ioannis Error-Related Potentials and Reinforcement Learning for Neuroadaptive Systems Proceedings Article In: BC25 : Bernstein Conference 2025,, BCCN Bernstein Network Computational Neuroscience & Neurotechnology , 2025. Abstract | Links | BibTeX | Schlagwörter: BCI, Machine Learning Saif-ur-Rehman, Muhammad; Ali, Omair; Klaes, Christian; Iossifidis, Ioannis In: Neurocomputing, S. 131370, 2025, ISSN: 0925-2312. Abstract | Links | BibTeX | Schlagwörter: BCI, Brain computer interface, Deep learning, Self organizing, Self-supervised machine learning, Spike Sorting Sziburis, Tim; Blex, Susanne; Glasmachers, Tobias; Iossifidis, Ioannis Hand Motion Catalog of Human Center-Out Transport Trajectories Measured Redundantly in 3D Task-Space Artikel In: Bd. 12, Nr. 1, S. 1293, 2025, ISSN: 2052-4463. Abstract | Links | BibTeX | Schlagwörter: BCI, Biomedical engineering, Motor control, Physiology Fidêncio, Aline Xavier; Grün, Felix; Klaes, Christian; Iossifidis, Ioannis Hybrid Brain-Computer Interface Using Error-Related Potential and Reinforcement Learning Artikel In: Frontiers in Human Neuroscience, Bd. 19, 2025, ISSN: 1662-5161. Abstract | Links | BibTeX | Schlagwörter: adaptive brain-computer interface, BCI, EEG, error-related potentials (ErrPs), Machine Learning, motor imagery (MI), reinforcement learning (RL) Fidêncio, Aline Xavier; Grün, Felix; Klaes, Christian; Iossifidis, Ioannis Error-Related Potential Driven Reinforcement Learning for Adaptive Brain-Computer Interfaces Artikel In: Arxiv, 2025. Abstract | Links | BibTeX | Schlagwörter: BCI, Computer Science - Human-Computer Interaction, Computer Science - Machine Learning, EEG, Quantitative Biology - Neurons and Cognition, Reinforcement learning Sziburis, Tim; Blex, Susanne; Glasmachers, Tobias; Iossifidis, Ioannis Deep-learning-based identification of individual motion characteristics from upper-limb trajectories towards disorder stage evaluation Proceedings Article In: Pons, Jose L.; Tornero, Jesus; Akay, Metin (Hrsg.): Converging Clinical and Engineering Research on Neurorehabilitation V - Proceedings of the 6th International Conference on Neurorehabilitation (ICNR2024), Springer International Publishing, La Granja, Spain, 2024. BibTeX | Schlagwörter: BCI, human arm motion, Motor control Fidencio, Aline Xavier; Klaes, Christian; Iossifidis, Ioannis Adaptive Brain-Computer Interfaces Based on Error-Related Potentials and Reinforcement Learning Proceedings Article In: BC24 : Computational Neuroscience & Neurotechnology Bernstein Conference 2024, BCCN Bernstein Network Computational Networkvphantom, 2024. Abstract | Links | BibTeX | Schlagwörter: BCI, Machine Learning Schmidt, Marie Dominique; Iossifidis, Ioannis Decoding Upper Limb Movements Proceedings Article In: BCCN Bernstein Network Computational Networkvphantom, 2024. Abstract | Links | BibTeX | Schlagwörter: BCI, Machine Learning, Muscle activity Fidêncio, Aline Xavier; Klaes, Christian; Iossifidis, Ioannis A Generic Error-Related Potential Classifier Based on Simulated Subjects Artikel In: Frontiers in Human Neuroscience, Bd. 18, S. 1390714, 2024, ISSN: 1662-5161. Abstract | Links | BibTeX | Schlagwörter: adaptive brain-machine (computer) interface, BCI, EEG, Error-related potential (ErrP), ErrP classifier, Generic decoder, Machine Learning, SEREEGA, Simulation Ali, Omair; Saif-ur-Rehman, Muhammad; Metzler, Marita; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian GET: A Generative EEG Transformer for Continuous Context-Based Neural Signals Artikel In: arXiv:2406.03115 [q-bio], 2024. Abstract | Links | BibTeX | Schlagwörter: BCI, EEG, Machine Learning, Quantitative Biology - Neurons and Cognition Ali, Omair; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian In: Computers in Biology and Medicine, S. 107649, 2023, ISSN: 0010-4825. Abstract | Links | BibTeX | Schlagwörter: BCI, Brain computer interface, Deep learning, EEG decoding, EMG decoding, Machine Learning Schmidt, Marie Dominique; Iossifidis, Ioannis The Link between Muscle Activity and Upper Limb Kinematics Proceedings Article In: BC23 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022, BCCN Bernstein Network Computational Network, 2023. Abstract | BibTeX | Schlagwörter: BCI, Machine Learning Fidencio, Aline Xavier; Klaes, Christian; Iossifidis, Ioannis Exploring Error-related Potentials in Adaptive Brain-Machine Interfaces: Challenges and Investigation of Occurrence and Detection Ratios Proceedings Article In: BC23 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022, BCCN Bernstein Network Computational Network, 2023. Abstract | BibTeX | Schlagwörter: BCI, EEG, Machine Learning Hussain, Muhammad Ayaz; Iossifidis, Ioannis In: arXiv:2309.04698 [cs.RO], 2023. Abstract | Links | BibTeX | Schlagwörter: BCI, Computer Science - Artificial Intelligence, Computer Science - Information Theory, Computer Science - Machine Learning, Exoskeleton, Robotics Schmidt, Marie D.; Glasmachers, Tobias; Iossifidis, Ioannis The Concepts of Muscle Activity Generation Driven by Upper Limb Kinematics Artikel In: BioMedical Engineering OnLine, Bd. 22, Nr. 1, S. 63, 2023, ISSN: 1475-925X. Abstract | Links | BibTeX | Schlagwörter: Artificial generated signal, BCI, Electromyography (EMG), Generative model, Inertial measurement unit (IMU), Machine Learning, Motion parameters, Muscle activity, Neural networks, transfer learning, Voluntary movement Saif-ur-Rehman, Muhammad; Ali, Omair; Klaes, Christian; Iossifidis, Ioannis In: arXiv:2304.01355 [cs, math, q-bio], 2023. Links | BibTeX | Schlagwörter: BCI, Machine Learning, Spike Sorting Lehmler, Stephan Johann; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis Deep transfer learning compared to subject-specific models for sEMG decoders Artikel In: Journal of Neural Engineering, Bd. 19, Nr. 5, 2022. Abstract | Links | BibTeX | Schlagwörter: BCI, Computational Complexity, Deep Transfer-Learning, Machine Learning, transfer learning Fidencio, Aline Xavier; Klaes, Christian; Iossifidis, Ioannis Closed-Loop Adaptation of Brain-Machine Interfaces Using Error-Related Potentials and Reinforcement Learning Proceedings Article In: BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022, BCCN Bernstein Network Computational Network, 2022. Links | BibTeX | Schlagwörter: BCI, Machine Learning Sziburis, Tim; Blex, Susanne; Iossifidis, Ioannis A Dataset of 3D Hand Transport Trajectories Determined by Inertial Measurements from a Single Sensor Proceedings Article In: BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022, BCCN Bernstein Network Computational Network, 2022. Links | BibTeX | Schlagwörter: BCI, Machine Learning Schmidt, Marie Dominique; Iossifidis, Ioannis Linking Muscle Activity and Motion Trajectory Proceedings Article In: BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022, BCCN Bernstein Network Computational Network, 2022. Links | BibTeX | Schlagwörter: BCI, Machine Learning Lehmler, Stephan Johann; Saif-ur-Rehman, Muhammad; Iossifidis, Ioannis Modeling Subject Specfic Surface EMG Features by Means of Deep Learning Proceedings Article In: BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022, BCCN Bernstein Network Computational Network, 2022. Links | BibTeX | Schlagwörter: BCI, Machine Learning Fidencio, Aline Xavier; Klaes, Christian; Iossifidis, Ioannis Error-Related Potentials in Reinforcement Learning-Based Brain-Machine Interfaces Artikel In: Frontiers in Human Neuroscience, Bd. 16, 2022. Abstract | Links | BibTeX | Schlagwörter: BCI, EEG, error-related potentials, Machine Learning, Reinforcement learning Ali, Omair; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian ConTraNet: A Single End-to-End Hybrid Network for EEG-based and EMG-based Human Machine Interfaces Artikel In: 2022. Abstract | Links | BibTeX | Schlagwörter: BCI, Machine Learning, neural processing, signal processing Doliwa, Sebastian; Erbeslöh, Andreas; Seidl, Karsten; Iossifidis, Ioannis Development of a Scalable Analog Front-End for Brain-Computer Interfaces Proceedings Article In: 17th International Conference on PhD Research in Microelectronics and Electronics, IEEE Prime 2022, Sardinia, Italy, 2022. BibTeX | Schlagwörter: BCI, Implantable BCI Ali, Omair; Saif-ur-Rehman, Muhammad; Dyck, Susanne; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian In: Nature Scientific Reports, Bd. 12, Ausg. 1, S. 4245, 2022, ISSN: 2045-2322. Abstract | Links | BibTeX | Schlagwörter: Adversarial NN, BCI, computer science, EEG, Machine Learning, Quantitative Biology, Quantitative Methods Schmidt, Marie Dominique; Glasmachers, Tobias; Iossifidis, Ioannis From Motion to Muscle Artikel In: arXiv: 2201.11501 [cs.LG], 2022. Links | BibTeX | Schlagwörter: BCI, Machine Learning, movement model, muscle signal generator, recurrent neural network Fidencio, Aline Xavier; Glasmachers, Tobias; Iossifidis, Ioannis Error-Related Potentials Detection with Dry- and Wet-Electrode EEG Proceedings Article In: FENS, Forum 2022, FENS, Federation of European Neuroscience Societies, 2022. Abstract | BibTeX | Schlagwörter: BCI, EEG, error-related potentials, Machine Learning Schmidt, Marie Dominique; Glasmachers, Tobias; Iossifidis, Ioannis Motion Intention Prediction Proceedings Article In: FENS, Forum 2022, FENS, Federation of European Neuroscience Societies, 2022. Abstract | BibTeX | Schlagwörter: BCI, Machine Learning, movement model, muscle signal generator, recurrent neural network Lehmler, Stephan Johann; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis Transfer-Learning for Patient Specific Model Re-Calibration: Application to sEMG-Classification Proceedings Article In: Bernstein Conferen, 2021. Links | BibTeX | Schlagwörter: BCI, Computational Complexity, Deep Transfer-Learning, Machine Learning, transfer learning Schmidt, Marie Dominique; Glasmachers, Tobias; Iossifidis, Ioannis Artificially Generated Muscle Signals Proceedings Article In: Bernstein Conference, 2021. Links | BibTeX | Schlagwörter: BCI, Machine Learning, movement model, muscle signal generator, recurrent neural network Fidencio, Aline Xavier; Glasmachers, Tobias; Klaes, Christian; Iossifidis, Ioannis Beyond Error Correction: Integration of Error-Related Potentials into Brain-Computer Interfaces for Improved Performance Proceedings Article In: Bernstein Conference, 2021. Links | BibTeX | Schlagwörter: BCI, error-related potentials, Machine Learning, Reinforcement learning Schmidt, Marie Dominique; Glasmachers, Tobias; Iossifidis, Ioannis Artificially Generated Muscle Signals Proceedings Article In: BC21 : Computational Neuroscience & Neurotechnology Bernstein Conference 2021, BCCN Bernstein Network Computational Network, 2021. Links | BibTeX | Schlagwörter: BCI, Machine Learning Lehmler, Stephan Johann; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis Transfer-Learning for Patient Specific Model Re-Calibration: Application to sEMG-Classification Proceedings Article In: BC21 : Computational Neuroscience & Neurotechnology Bernstein Conference 2021, BCCN Bernstein Network Computational Network, 2021. Links | BibTeX | Schlagwörter: BCI, Machine Learning Sziburis, Tim; Blex, Susanne; Glasmachers, Tobias; Rano, Inaki; Iossifidis, Ioannis Modelling the Generation of Human Upper-Limb Reaching Trajectories: An Extended Behavioural Attractor Dynamics Approach Proceedings Article In: BC21 : Computational Neuroscience & Neurotechnology Bernstein Conference 2021, BCCN Bernstein Network Computational Network, 2021. Links | BibTeX | Schlagwörter: BCI, Machine Learning, movement model Ali, Omair; Saif-ur-Rehman, Muhammad; Dyck, Susanne; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian Anchored-STFT and GNAA: An Extension of STFT in Conjunction with an Adversarial Data Augmentation Technique for the Decoding of Neural Signals Artikel In: arXiv:2011.14694 [cs, q-bio], 2021. Abstract | BibTeX | Schlagwörter: BCI, Machine Learning, Quantitative Biology, Quantitative Methods Lehmler, Stephan Johann; Saif-ur-Rehman, Muhammad; Glasmachers, Tobias; Iossifidis, Ioannis Deep Transfer-Learning for patient specific model re-calibration: Application to sEMG-Classification Artikel In: 2021. Links | BibTeX | Schlagwörter: BCI, Computational Complexity, Deep Transfer-Learning, Machine Learning Saif-ur-Rehman, Muhammad; Ali, Omair; Dyck, Susanne; Lienkämper, Robin; Metzler, Marita; Parpaley, Yaroslav; Wellmer, Jörg; Liu, Charles; Lee, Brian; Kellis, Spencer; Andersen, Richard; Iossifidis, Ioannis; Glasmachers, Tobias; Klaes, Christian SpikeDeep-Classifier: A deep-learning based fully automatic offline spike sorting algorithm Artikel In: Journal of Neural Engineering, 2020. Abstract | Links | BibTeX | Schlagwörter: BCI, CNN, Machine Learning, Spike Sorting Ali, Omair; Saif-ur-Rehman, Muhammad; Dyck, Susanne; Glasmachers, Tobias; Iossifidis, Ioannis; Klaes, Christian Improving the performance of EEG decoding using anchored-STFT in conjunction with gradient norm adversarial augmentation Artikel In: arXiv preprint arXiv:2011.14694, 2020. BibTeX | Schlagwörter: Adversarial NN, BCI, EEG, Machine Learning Saif-ur-Rehman, Muhammad; Lienkämper, Robin; Dyck, Susanne; Rayana, A; Parpaley, Y; Wllner, J; Liu, Charles; Lee, Brian; Kellis, Spencer; Manahan-Vaughn, D; Güntürkün, O; Andersen, Richard; Iossifidis, Ioannis; Glasmachers, Tobias; Klaes, Christian Universal SpikeDeeptector Sonstige 2019. Abstract | BibTeX | Schlagwörter: BCI, CNN, Machine Learning, Spike Detection, Spike Sorting Saif-ur-Rehman, Muhammad; Lienkämper, Robin; Parpaley, Yaroslav; Wellmer, Jörg; Liu, Charles; Lee, Brian; Kellis, Spencer; Andersen, Richard; Iossifidis, Ioannis; Glasmachers, Tobias; Klaes, Christian SpikeDeeptector: a deep-learning based method for detection of neural spiking activity Artikel In: Journal of Neural Engineering, Bd. 16, Nr. 5, S. 056003, 2019. Abstract | Links | BibTeX | Schlagwörter: BCI, CNN, Data Reduction, Machine Learning, Spike Sorting Iossifidis, Ioannis; Klaes, Christian Low dimensional representation of human arm movement for efficient neuroprosthetic control by individuals with tetraplegia Sonstige 2017. Abstract | BibTeX | Schlagwörter: BCI, Dynamical systems, movement model, neuroprosthetic, Robotics Klaes, Christian; Iossifidis, Ioannis Low dimensional representation of human arm movement for efficient neuroprosthetic control by individuals with tetraplegia Konferenz SfN Meeting 2017, 2017. BibTeX | Schlagwörter: BCI, Dynamical systems, movement model, neuroprosthetic, Robotics2026
@article{schmidtInsightsMotorControl2026,
title = {Insights into Motor Control: Predict Muscle Activity from Upper Limb Kinematics with LSTM Networks},
author = {Marie D. Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
editor = {Nature Publishing Group},
url = {https://www.nature.com/articles/s41598-025-33696-y},
doi = {10.1038/s41598-025-33696-y},
issn = {2045-2322},
year = {2026},
date = {2026-01-05},
urldate = {2026-01-05},
journal = {Nature Scientific Reports},
publisher = {Nature Publishing Group},
abstract = {This study explores the relationship between upper limb kinematics and corresponding muscle activity, aiming to understand how predictive models can approximate motor control. We employ a Long Short-Term Memory (LSTM) network trained on kinematic end effector data to estimate muscle activity for eight muscles. The model exhibits strong predictive accuracy for new repetitions of known movements and generalizes to unseen movements, suggesting it captures underlying biomechanical principles rather than merely memorizing patterns. This generalization is particularly valuable for applications in rehabilitation and human-machine interaction, as it reduces the need for exhaustive datasets. To further investigate movement representation and learning, we analyze the impact of motion segmentation, hypothesizing that breaking movements into simpler components may improve model performance. Additionally, we explore the role of the swivel angle in reducing redundancy in arm kinematics. Another key focus is the effect of training data complexity on generalization. Specifically, we assess whether training on a diverse set of movements leads to better performance than specializing in either simple, single-joint movements or complex, multi-joint movements. The study is based on an experimental setup involving 23 distinct upper limb movements performed by five subjects. Our findings provide insights into the interplay between kinematics and muscle activity, contributing to motor control research and advancing neural network-based movement prediction.},
keywords = {BCI, Computational biology and bioinformatics, Motor control, Neuroscience},
pubstate = {published},
tppubtype = {article}
}
2025
@inproceedings{schmidtPredictingUpperLimb2025,
title = {Predicting Upper Limb Muscle Activity from Kinematics Using LSTM Networks},
author = {Marie Dominique Schmidt and Ioannis Iossifidis},
url = {https://abstracts.g-node.org/conference/BC25/abstracts#/uuid/a8fa799f-bf8e-4192-8ba9-3b1e3ad078ea},
year = {2025},
date = {2025-10-01},
urldate = {2025-10-01},
booktitle = {Bernstein Conference 2025},
publisher = {BCCN Bernstein Network Computational Neuroscience & Neurotechnology },
abstract = {The upper limbs are essential for performing everyday tasks that require a wide range of motion and precise coordination. Planning and timing are crucial to achieve coordinated movement. Sensory information about the target and current body state is critical, as is the integration of prior experience represented by prelearned inverse dynamics that generate the associated muscle activity. We propose a generative model that uses a recurrent neural network to predict upper limb muscle activity during various simple and complex everyday movements. By identifying movement primitives within the signal, our model enables the decomposition of these movements into a fundamental set, facilitating the reconstruction of muscle activity patterns. Our approach has implications for the fundamental understanding of movement control and the rehabilitation of neuromuscular disorders with myoelectric prosthetics and functional electrical stimulation.},
keywords = {BCI},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{xavierfidencioErrorrelatedPotentialsReinforcement2025,
title = {Error-Related Potentials and Reinforcement Learning for Neuroadaptive Systems},
author = {Aline Xavier Fidencio and Ioannis Iossifidis},
url = {https://abstracts.g-node.org/conference/BC25/abstracts#/uuid/84a56344-585c-4d7c-992d-1f34489770f5},
year = {2025},
date = {2025-10-01},
urldate = {2025-10-01},
booktitle = {BC25 : Bernstein Conference 2025,},
publisher = {BCCN Bernstein Network Computational Neuroscience & Neurotechnology },
abstract = {Error-related potentials (ErrPs) represent the neural signature of error processing in the brain and numerous studies have demonstrated their reliable detection using non-invasive techniques such as electroencephalography (EEG). Over recent decades, the brain-computer interface (BCI) community has shown growing interest in leveraging these intrinsic feedback signals to enhance system performance. However, the effective use of ErrPs in a closed-loop setup crucially depends on accurate single-trial detection, which is typically achieved using a subject-specific classifier (or decoder) trained on samples recorded during extensive calibration sessions before the BCI system can be deployed. In our research, we explore the potential of simulated EEG data for training a truly generic ErrP classifier. Utilizing the SEREEGA simulator, we demonstrate that EEG data can be generated in a cost-effective manner, allowing for controlled and systematic variations in data distribution to accommodate uncertainties in ErrP generation. A classifier trained solely on the generated data exhibits promising generalization capabilities across different datasets and performs comparably to a leave-one-subject-out approach trained on real data (Xavier Fidêncio et al., 2024). In our experiments, we deliberately provoked ErrPs when the BCI misinterpreted the user's intention, resulting in incorrect actions. Subjects engaged in a game controlled via keyboard and/or motor imagery (imagining hand movements), with EEG data recorded using various EEG systems for comparison. Considering the challenges in obtaining clear ErrP signals for all subjects and the limitations identified in existing literature (Xavier Fidêncio et al., 2022), we hypothesize whether a measurable error signal is consistently generated at the scalp level when subjects encounter erroneous conditions, and how this influences closed-loop setups that incorporate ErrPs for improved BCI performance. To address these questions, we assess the effects of the occurrence-to-detection ratio of ErrPs in the classification pipeline using simulated data and explore the impact of error misclassification rates in an ErrP-based learning framework, which employs reinforcement learning to enhance BCI performance.},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@article{saif-ur-rehmanAdaptiveSpikeDeepclassifierSelforganizing2025,
title = {Adaptive SpikeDeep-classifier: Self-organizing and Self-Supervised Machine Learning Algorithm for Online Spike Sorting},
author = {Muhammad Saif-ur-Rehman and Omair Ali and Christian Klaes and Ioannis Iossifidis},
editor = {Elsevier},
url = {https://www.sciencedirect.com/science/article/pii/S0925231225020429},
doi = {10.1016/j.neucom.2025.131370},
issn = {0925-2312},
year = {2025},
date = {2025-09-04},
urldate = {2025-09-04},
journal = {Neurocomputing},
pages = {131370},
abstract = {Objective. Invasive brain-computer interface (BCI) research is progressing towards the realization of the motor skills rehabilitation of severely disabled patients in the real world. The size of invasive micro-electrode arrays and the selection of an efficient online spike sorting algorithm (performing spike sorting at run time) are two key factors that play pivotal roles in the successful decoding of the user’s intentions. The process of spike sorting includes the selection of channels that record the spike activity (SA) and determines the SA of different sources (neurons), on selected channels individually. The neural data recorded with dense micro-electrode arrays is time-varying and often contaminated with non-stationary noise. Unfortunately, current state-of-the-art spike sorting algorithms are incapable of handling the massively increasing amount of time-varying data resulting from the dense microelectrode arrays, which makes the spike sorting one of the fragile components of the online BCI decoding framework. Approach. This study proposed an adaptive and self-organized algorithm for online spike sorting, named as Adaptive SpikeDeep-Classifier (Ada-SpikeDeepClassifier). Our algorithm uses SpikeDeeptector for the channel selection, an adaptive background activity rejector (Ada-BAR) for discarding the background events, and an adaptive spike deep-classifier (Ada-SpikeDeepClassifier) for classifying the SA of different neural units. The process of spike sorting is accomplished by concatenating SpikeDeeptector, Ada-BAR and Ada-SpikeDeepclassifier. Results. The proposed algorithm is evaluated on two different categories of data: a human data-set recorded in our lab, and a publicly available simulated data-set to avoid subjective biases and labeling errors. The proposed Ada-SpikeDeepClassifier outperformed our previously published SpikeDeep-Classifier and eight conventional spike sorting algorithms and produce comparable results to state of the art deep learning based algorithms. Significance. To the best of our knowledge, the proposed algorithm is the first spike sorting algorithm that autonomously adapts to the shift in the distribution of noise and SA data and perform spike sorting without human interventions in various kinds of experimental settings. In addition, the proposed algorithm builds upon artificial neural networks, which makes it an ideal candidate for being embedded on neuromorphic chips that are also suitable for wearable invasive BCI.},
keywords = {BCI, Brain computer interface, Deep learning, Self organizing, Self-supervised machine learning, Spike Sorting},
pubstate = {published},
tppubtype = {article}
}
@article{sziburisHandMotionCatalog2025,
title = {Hand Motion Catalog of Human Center-Out Transport Trajectories Measured Redundantly in 3D Task-Space},
author = {Tim Sziburis and Susanne Blex and Tobias Glasmachers and Ioannis Iossifidis},
editor = {Nature},
url = {https://www.nature.com/articles/s41597-025-05576-7},
doi = {10.1038/s41597-025-05576-7},
issn = {2052-4463},
year = {2025},
date = {2025-07-24},
urldate = {2025-07-24},
volume = {12},
number = {1},
pages = {1293},
publisher = {Nature Publishing Group},
abstract = {Motion modeling and variability analysis bear the potential to identify movement pathology but require profound data. We introduce a systematic dataset of 3D center-out task-space trajectories of human hand transport movements in a standardized setting. This set-up is characterized by reproducibility, leading to reliable transferability to various locations. The transport tasks consist of grasping a cylindrical object from a unified start position and transporting it to one of nine target locations in unconstrained operational space. The measurement procedure is automatized to record ten trials per target location and participant. The dataset comprises 90 movement trajectories for each hand of 31 participants without known movement disorders (21 to 78 years), resulting in 5580 trials. In addition, handedness is determined using the EHI. Data are recorded redundantly and synchronously by an optical tracking system and a single IMU sensor. Unlike the stationary capturing system, the IMU can be considered a portable, low-cost, and energy-efficient alternative to be implemented on embedded systems, for example in medical evaluation scenarios.},
keywords = {BCI, Biomedical engineering, Motor control, Physiology},
pubstate = {published},
tppubtype = {article}
}
@article{xavierfidencioHybridBraincomputerInterface2025,
title = {Hybrid Brain-Computer Interface Using Error-Related Potential and Reinforcement Learning},
author = {Aline Xavier Fidêncio and Felix Grün and Christian Klaes and Ioannis Iossifidis},
editor = {Frontiers},
url = {https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2025.1569411/full},
doi = {10.3389/fnhum.2025.1569411},
issn = {1662-5161},
year = {2025},
date = {2025-06-04},
urldate = {2025-06-04},
journal = {Frontiers in Human Neuroscience},
volume = {19},
publisher = {Frontiers},
abstract = {Brain-computer interfaces (BCIs) offer alternative communication methods for individuals with motor disabilities, aiming to improve their quality of life through external device control. However, non-invasive BCIs using electroencephalography (EEG) often suffer from performance limitations due to non-stationarities arising from changes in mental state or device characteristics. Addressing these challenges motivates the development of adaptive systems capable of real-time adjustment. This study investigates a novel approach for creating an adaptive, error-related potential (ErrP)-based BCI using reinforcement learning (RL) to dynamically adapt to EEG signal variations. The framework was validated through experiments on a publicly available motor imagery dataset and a novel fast-paced protocol designed to enhance user engagement. Results showed that RL agents effectively learned control policies from user interactions, maintaining robust performance across datasets. However, findings from the game-based protocol revealed that fast-paced motor imagery tasks were ineffective for most participants, highlighting critical challenges in real-time BCI task design. Overall, the results demonstrate the potential of RL for enhancing BCI adaptability while identifying practical constraints in task complexity and user responsiveness.},
keywords = {adaptive brain-computer interface, BCI, EEG, error-related potentials (ErrPs), Machine Learning, motor imagery (MI), reinforcement learning (RL)},
pubstate = {published},
tppubtype = {article}
}
@article{fidencioErrorrelatedPotentialDriven2025,
title = {Error-Related Potential Driven Reinforcement Learning for Adaptive Brain-Computer Interfaces},
author = {Aline Xavier Fidêncio and Felix Grün and Christian Klaes and Ioannis Iossifidis},
url = {http://arxiv.org/abs/2502.18594},
doi = {10.48550/arXiv.2502.18594},
year = {2025},
date = {2025-02-25},
urldate = {2025-02-25},
journal = {Arxiv},
abstract = {Brain-computer interfaces (BCIs) provide alternative communication methods for individuals with motor disabilities by allowing control and interaction with external devices. Non-invasive BCIs, especially those using electroencephalography (EEG), are practical and safe for various applications. However, their performance is often hindered by EEG non-stationarities, caused by changing mental states or device characteristics like electrode impedance. This challenge has spurred research into adaptive BCIs that can handle such variations. In recent years, interest has grown in using error-related potentials (ErrPs) to enhance BCI performance. ErrPs, neural responses to errors, can be detected non-invasively and have been integrated into different BCI paradigms to improve performance through error correction or adaptation. This research introduces a novel adaptive ErrP-based BCI approach using reinforcement learning (RL). We demonstrate the feasibility of an RL-driven adaptive framework incorporating ErrPs and motor imagery. Utilizing two RL agents, the framework adapts dynamically to EEG non-stationarities. Validation was conducted using a publicly available motor imagery dataset and a fast-paced game designed to boost user engagement. Results show the framework's promise, with RL agents learning control policies from user interactions and achieving robust performance across datasets. However, a critical insight from the game-based protocol revealed that motor imagery in a high-speed interaction paradigm was largely ineffective for participants, highlighting task design limitations in real-time BCI applications. These findings underscore the potential of RL for adaptive BCIs while pointing out practical constraints related to task complexity and user responsiveness.},
keywords = {BCI, Computer Science - Human-Computer Interaction, Computer Science - Machine Learning, EEG, Quantitative Biology - Neurons and Cognition, Reinforcement learning},
pubstate = {published},
tppubtype = {article}
}
2024
@inproceedings{icnr2024,
title = {Deep-learning-based identification of individual motion characteristics from upper-limb trajectories towards disorder stage evaluation},
author = {Tim Sziburis and Susanne Blex and Tobias Glasmachers and Ioannis Iossifidis},
editor = {Jose L. Pons and Jesus Tornero and Metin Akay},
year = {2024},
date = {2024-11-30},
urldate = {2024-11-30},
booktitle = {Converging Clinical and Engineering Research on Neurorehabilitation V - Proceedings of the 6th International Conference on Neurorehabilitation (ICNR2024)},
publisher = {Springer International Publishing},
address = {La Granja, Spain},
series = {Biosystems and Biorobotics},
keywords = {BCI, human arm motion, Motor control},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{AdaptiveBraincomputerInterfaces2024,
title = {Adaptive Brain-Computer Interfaces Based on Error-Related Potentials and Reinforcement Learning},
author = {Aline Xavier Fidencio and Christian Klaes and Ioannis Iossifidis},
url = {https://abstracts.g-node.org/conference/BC24/abstracts#/uuid/03d3dd16-4c50-43d8-b878-abcfa7857386},
year = {2024},
date = {2024-09-18},
urldate = {2024-09-24},
booktitle = {BC24 : Computational Neuroscience & Neurotechnology Bernstein Conference 2024},
publisher = {BCCN Bernstein Network Computational Networkvphantom},
abstract = {Error-related potentials (ErrPs) represent the neural signature of error processing in the brain and numerous studies have demonstrated their reliable detection using non-invasive techniques such as electroencephalography (EEG). Over recent decades, the brain-computer interface (BCI) community has shown growing interest in leveraging these intrinsic feedback signals to enhance system performance. However, the effective use of ErrPs in a closed-loop setup crucially depends on accurate single-trial detection, which is typically achieved using a subject-specific classifier (or decoder) trained on samples recorded during extensive calibration sessions before the BCI system can be deployed. In our research, we explore the potential of simulated EEG data for training a truly generic ErrP classifier. Utilizing the SEREEGA simulator, we demonstrate that EEG data can be generated in a cost-effective manner, allowing for controlled and systematic variations in data distribution to accommodate uncertainties in ErrP generation. A classifier trained solely on the generated data exhibits promising generalization capabilities across different datasets and performs comparably to a leave-one-subject-out approach trained on real data (Xavier Fidêncio et al., 2024). In our experiments, we deliberately provoked ErrPs when the BCI misinterpreted the user's intention, resulting in incorrect actions. Subjects engaged in a game controlled via keyboard and/or motor imagery (imagining hand movements), with EEG data recorded using various EEG systems for comparison. Considering the challenges in obtaining clear ErrP signals for all subjects and the limitations identified in existing literature (Xavier Fidêncio et al., 2022), we hypothesize whether a measurable error signal is consistently generated at the scalp level when subjects encounter erroneous conditions, and how this influences closed-loop setups that incorporate ErrPs for improved BCI performance. To address these questions, we assess the effects of the occurrence-to-detection ratio of ErrPs in the classification pipeline using simulated data and explore the impact of error misclassification rates in an ErrP-based learning framework, which employs reinforcement learning to enhance BCI performance.},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{DecodingUpperLimb2024,
title = {Decoding Upper Limb Movements},
author = {Marie Dominique Schmidt and Ioannis Iossifidis},
url = {https://abstracts.g-node.org/conference/BC24/abstracts#/uuid/4725140f-ce7c-4ac5-b694-c627ceeb8d98},
year = {2024},
date = {2024-09-18},
urldate = {2024-09-24},
publisher = {BCCN Bernstein Network Computational Networkvphantom},
abstract = {The upper limbs are essential for performing everyday tasks that require a wide range of motion and precise coordination. Planning and timing are crucial to achieve coordinated movement. Sensory information about the target and current body state is critical, as is the integration of prior experience represented by prelearned inverse dynamics that generate the associated muscle activity. We propose a generative model that uses a recurrent neural network to predict upper limb muscle activity during various simple and complex everyday movements. By identifying movement primitives within the signal, our model enables the decomposition of these movements into a fundamental set, facilitating the reconstruction of muscle activity patterns. Our approach has implications for the fundamental understanding of movement control and the rehabilitation of neuromuscular disorders with myoelectric prosthetics and functional electrical stimulation.},
keywords = {BCI, Machine Learning, Muscle activity},
pubstate = {published},
tppubtype = {inproceedings}
}
@article{xavierfidencioGenericErrorrelatedPotential2024,
title = {A Generic Error-Related Potential Classifier Based on Simulated Subjects},
author = {Aline Xavier Fidêncio and Christian Klaes and Ioannis Iossifidis},
editor = {Frontiers Media SA},
url = {https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2024.1390714/full},
doi = {10.3389/fnhum.2024.1390714},
issn = {1662-5161},
year = {2024},
date = {2024-07-19},
urldate = {2024-07-19},
journal = {Frontiers in Human Neuroscience},
volume = {18},
pages = {1390714},
publisher = {Frontiers},
abstract = {$<$p$>$Error-related potentials (ErrPs) are brain signals known to be generated as a reaction to erroneous events. Several works have shown that not only self-made errors but also mistakes generated by external agents can elicit such event-related potentials. The possibility of reliably measuring ErrPs through non-invasive techniques has increased the interest in the brain-computer interface (BCI) community in using such signals to improve performance, for example, by performing error correction. Extensive calibration sessions are typically necessary to gather sufficient trials for training subject-specific ErrP classifiers. This procedure is not only time-consuming but also boresome for participants. In this paper, we explore the effectiveness of ErrPs in closed-loop systems, emphasizing their dependency on precise single-trial classification. To guarantee the presence of an ErrPs signal in the data we employ and to ensure that the parameters defining ErrPs are systematically varied, we utilize the open-source toolbox SEREEGA for data simulation. We generated training instances and evaluated the performance of the generic classifier on both simulated and real-world datasets, proposing a promising alternative to conventional calibration techniques. Results show that a generic support vector machine classifier reaches balanced accuracies of 72.9%, 62.7%, 71.0%, and 70.8% on each validation dataset. While performing similarly to a leave-one-subject-out approach for error class detection, the proposed classifier shows promising generalization across different datasets and subjects without further adaptation. Moreover, by utilizing SEREEGA, we can systematically adjust parameters to accommodate the variability in the ErrP, facilitating the systematic validation of closed-loop setups. Furthermore, our objective is to develop a universal ErrP classifier that captures the signal's variability, enabling it to determine the presence or absence of an ErrP in real EEG data.$<$/p$>$},
keywords = {adaptive brain-machine (computer) interface, BCI, EEG, Error-related potential (ErrP), ErrP classifier, Generic decoder, Machine Learning, SEREEGA, Simulation},
pubstate = {published},
tppubtype = {article}
}
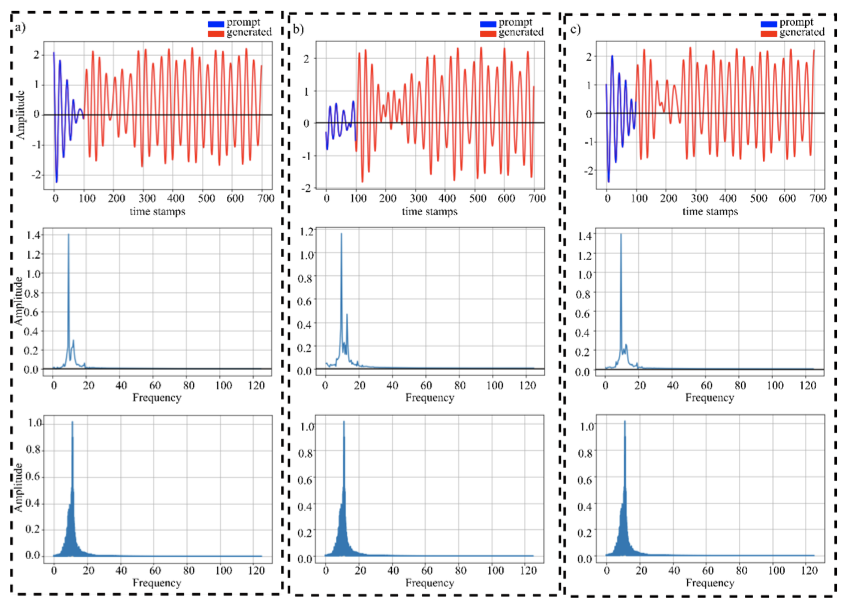
@article{aliGETGenerativeEEG2024,
title = {GET: A Generative EEG Transformer for Continuous Context-Based Neural Signals},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Marita Metzler and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {http://arxiv.org/abs/2406.03115},
doi = {10.48550/arXiv.2406.03115},
year = {2024},
date = {2024-06-09},
urldate = {2024-06-09},
journal = {arXiv:2406.03115 [q-bio]},
abstract = {Generating continuous electroencephalography (EEG) signals through advanced artificial neural networks presents a novel opportunity to enhance brain-computer interface (BCI) technology. This capability has the potential to significantly enhance applications ranging from simulating dynamic brain activity and data augmentation to improving real-time epilepsy detection and BCI inference. By harnessing generative transformer neural networks, specifically designed for EEG signal generation, we can revolutionize the interpretation and interaction with neural data. Generative AI has demonstrated significant success across various domains, from natural language processing (NLP) and computer vision to content creation in visual arts and music. It distinguishes itself by using large-scale datasets to construct context windows during pre-training, a technique that has proven particularly effective in NLP, where models are fine-tuned for specific downstream tasks after extensive foundational training. However, the application of generative AI in the field of BCIs, particularly through the development of continuous, context-rich neural signal generators, has been limited. To address this, we introduce the Generative EEG Transformer (GET), a model leveraging transformer architecture tailored for EEG data. The GET model is pre-trained on diverse EEG datasets, including motor imagery and alpha wave datasets, enabling it to produce high-fidelity neural signals that maintain contextual integrity. Our empirical findings indicate that GET not only faithfully reproduces the frequency spectrum of the training data and input prompts but also robustly generates continuous neural signals. By adopting the successful training strategies of the NLP domain for BCIs, the GET sets a new standard for the development and application of neural signal generation technologies.},
keywords = {BCI, EEG, Machine Learning, Quantitative Biology - Neurons and Cognition},
pubstate = {published},
tppubtype = {article}
}
2023
@article{aliConTraNetHybridNetwork2023,
title = {ConTraNet: A Hybrid Network for Improving the Classification of EEG and EMG Signals with Limited Training Data},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {https://www.sciencedirect.com/science/article/pii/S0010482523011149},
doi = {10.1016/j.compbiomed.2023.107649},
issn = {0010-4825},
year = {2023},
date = {2023-11-02},
urldate = {2023-11-02},
journal = {Computers in Biology and Medicine},
pages = {107649},
abstract = {Objective Bio-Signals such as electroencephalography (EEG) and electromyography (EMG) are widely used for the rehabilitation of physically disabled people and for the characterization of cognitive impairments. Successful decoding of these bio-signals is however non-trivial because of the time-varying and non-stationary characteristics. Furthermore, existence of short- and long-range dependencies in these time-series signal makes the decoding even more challenging. State-of-the-art studies proposed Convolutional Neural Networks (CNNs) based architectures for the classification of these bio-signals, which are proven useful to learn spatial representations. However, CNNs because of the fixed size convolutional kernels and shared weights pay only uniform attention and are also suboptimal in learning short-long term dependencies, simultaneously, which could be pivotal in decoding EEG and EMG signals. Therefore, it is important to address these limitations of CNNs. To learn short- and long-range dependencies simultaneously and to pay more attention to more relevant part of the input signal, Transformer neural network-based architectures can play a significant role. Nonetheless, it requires a large corpus of training data. However, EEG and EMG decoding studies produce limited amount of the data. Therefore, using standalone transformers neural networks produce ordinary results. In this study, we ask a question whether we can fix the limitations of CNN and transformer neural networks and provide a robust and generalized model that can simultaneously learn spatial patterns, long-short term dependencies, pay variable amount of attention to time-varying non-stationary input signal with limited training data. Approach In this work, we introduce a novel single hybrid model called ConTraNet, which is based on CNN and Transformer architectures that contains the strengths of both CNN and Transformer neural networks. ConTraNet uses a CNN block to introduce inductive bias in the model and learn local dependencies, whereas the Transformer block uses the self-attention mechanism to learn the short- and long-range or global dependencies in the signal and learn to pay different attention to different parts of the signals. Main results We evaluated and compared the ConTraNet with state-of-the-art methods on four publicly available datasets (BCI Competition IV dataset 2b, Physionet MI-EEG dataset, Mendeley sEMG dataset, Mendeley sEMG V1 dataset) which belong to EEG-HMI and EMG-HMI paradigms. ConTraNet outperformed its counterparts in all the different category tasks (2-class, 3-class, 4-class, 7-class, and 10-class decoding tasks). Significance With limited training data ConTraNet significantly improves classification performance on four publicly available datasets for 2, 3, 4, 7, and 10-classes compared to its counterparts.},
keywords = {BCI, Brain computer interface, Deep learning, EEG decoding, EMG decoding, Machine Learning},
pubstate = {published},
tppubtype = {article}
}
@inproceedings{schmidtLinkMuscleActivity2023,
title = {The Link between Muscle Activity and Upper Limb Kinematics},
author = {Marie Dominique Schmidt and Ioannis Iossifidis},
year = {2023},
date = {2023-09-15},
urldate = {2023-09-15},
booktitle = {BC23 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022},
publisher = {BCCN Bernstein Network Computational Network},
abstract = {The upper limbs are crucial in performing daily tasks that require strength, a wide range of motion, and precision. To achieve coordinated motion, planning and timing are critical. Sensory information about the target and the current body state is essential, as well as integrating past experiences, represented by pre-learned inverse dynamics that generate associated muscle activity. We propose a generative model that predicts upper limb muscle activity from a variety of simple and complex everyday motions by means of a recurrent neural network. The model shows promising results, with a good fit for different subjects and abstracts well for new motions. We handle the high inter-subject variation in muscle activity using a transfer learning approach, resulting in a good fit for new subjects. Our approach has implications for fundamental movement control understanding and the rehabilitation of neuromuscular diseases using myoelectric prostheses and functional electrical stimulation. Our model can efficiently predict both muscle activity and motion trajectory, which can assist in developing more effective rehabilitation techniques.},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{xavierfidencioExploringErrorrelatedPotentials2023,
title = {Exploring Error-related Potentials in Adaptive Brain-Machine Interfaces: Challenges and Investigation of Occurrence and Detection Ratios},
author = {Aline Xavier Fidencio and Christian Klaes and Ioannis Iossifidis},
year = {2023},
date = {2023-09-15},
urldate = {2023-09-15},
booktitle = {BC23 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022},
publisher = {BCCN Bernstein Network Computational Network},
abstract = {Non-invasive techniques like EEG can record error-related potentials (ErrPs), neural signals associated with error processing and awareness. ErrPs are generated in response to self-made and external errors, including those produced by the BMI. Since ErrPs are implicitly elicited and don’t add extra workload for the subject, they serve as a natural and intrinsic feedback source for developing adaptive BMIs. In our study, we assess the occurrence of interaction ErrPs in an adaptive BMI that combines ErrPs and reinforcement learning. We intentionally provoke ErrPs when the BMI misinterprets the user’s intention and performs an incorrect action. Subjects participated in a game controlled by a keyboard and/or motor imagery (imagining hand movements), and EEG data were recorded using an eight-electrode gel-based EEG system. Results reveal that obtaining a distinct ErrPs signal for each subject is more challenging than anticipated. Current practices report the ErrP in terms of over all subjects and trials difference grand average (error minus correct). This approach has, however, the limitation of masking the inter-trial and subject variability, which are relevant for the online single-trial detection of such signals. Moreover, the reported ErrPs waveshape exhibit differences in terms of components observed, as well as their respective latencies, even when very similar tasks are used. Consequently, we conducted additional individualized data analysis to gain deeper insights into the single-trial nature of the ErrPs. As a result, we determined the need for a better understanding and further investigation of how effectively the ErrPs waveforms generalize across subjects, tasks, experimental protocols, and feedback modalities. Given the challenges in obtaining a clear signal for all subjects and the limitations found in existing literature (Xavier Fidêncio et al., 2022), we hypothesize whether an error signal measurable at the scalp level is consistently generated when subjects encounter erroneous conditions. To address this question, we will assess the occurrence-to-detection ratio of ErrPs using invasive and non-invasive recording techniques, examining how uncertainties regarding error generation in the brain impact the learning pipeline.},
keywords = {BCI, EEG, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}

@article{ayazhussainAdvancementsUpperBody2023,
title = {Advancements in Upper Body Exoskeleton: Implementing Active Gravity Compensation with a Feedforward Controller},
author = {Muhammad Ayaz Hussain and Ioannis Iossifidis},
url = {https://doi.org/10.48550/arXiv.2309.04698},
doi = {10.48550/arXiv.2309.04698},
year = {2023},
date = {2023-09-09},
urldate = {2023-09-09},
journal = {arXiv:2309.04698 [cs.RO]},
abstract = {In this study, we present a feedforward control system designed for active gravity compensation on an upper body exoskeleton. The system utilizes only positional data from internal motor sensors to calculate torque, employing analytical control equations based on Newton-Euler Inverse Dynamics. Compared to feedback control systems, the feedforward approach offers several advantages. It eliminates the need for external torque sensors, resulting in reduced hardware complexity and weight. Moreover, the feedforward control exhibits a more proactive response, leading to enhanced performance. The exoskeleton used in the experiments is lightweight and comprises 4 Degrees of Freedom, closely mimicking human upper body kinematics and three-dimensional range of motion. We conducted tests on both hardware and simulations of the exoskeleton, demonstrating stable performance. The system maintained its position over an extended period, exhibiting minimal friction and avoiding undesired slewing.},
keywords = {BCI, Computer Science - Artificial Intelligence, Computer Science - Information Theory, Computer Science - Machine Learning, Exoskeleton, Robotics},
pubstate = {published},
tppubtype = {article}
}
@article{schmidtConceptsMuscleActivity2023,
title = {The Concepts of Muscle Activity Generation Driven by Upper Limb Kinematics},
author = {Marie D. Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
url = {https://doi.org/10.1186/s12938-023-01116-9},
doi = {10.1186/s12938-023-01116-9},
issn = {1475-925X},
year = {2023},
date = {2023-06-24},
urldate = {2023-06-24},
journal = {BioMedical Engineering OnLine},
volume = {22},
number = {1},
pages = {63},
abstract = {The underlying motivation of this work is to demonstrate that artificial muscle activity of known and unknown motion can be generated based on motion parameters, such as angular position, acceleration, and velocity of each joint (or the end-effector instead), which are similarly represented in our brains. This model is motivated by the known motion planning process in the central nervous system. That process incorporates the current body state from sensory systems and previous experiences, which might be represented as pre-learned inverse dynamics that generate associated muscle activity.},
keywords = {Artificial generated signal, BCI, Electromyography (EMG), Generative model, Inertial measurement unit (IMU), Machine Learning, Motion parameters, Muscle activity, Neural networks, transfer learning, Voluntary movement},
pubstate = {published},
tppubtype = {article}
}
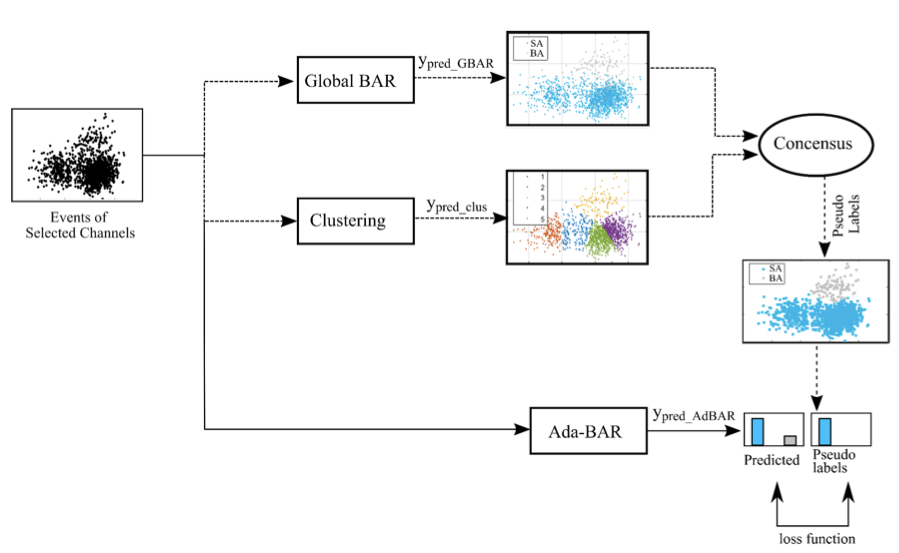
@article{saifurrehman2023adaptive,
title = {Adaptive SpikeDeep-Classifier: Self-organizing and self-supervised machine learning algorithm for online spike sorting},
author = {Muhammad Saif-ur-Rehman and Omair Ali and Christian Klaes and Ioannis Iossifidis},
doi = {10.48550/arXiv.2304.01355},
year = {2023},
date = {2023-05-02},
urldate = {2023-05-02},
journal = {arXiv:2304.01355 [cs, math, q-bio]},
keywords = {BCI, Machine Learning, Spike Sorting},
pubstate = {published},
tppubtype = {article}
}
2022
@article{lehmlerTransferLearningPatientSpecific2021bb,
title = {Deep transfer learning compared to subject-specific models for sEMG decoders},
author = {Stephan Johann Lehmler and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis},
editor = {{IOP Publishing},
url = {https://dx.doi.org/10.1088/1741-2552/ac9860},
doi = {10.1088/1741-2552/ac9860},
year = {2022},
date = {2022-10-28},
urldate = {2022-10-28},
journal = {Journal of Neural Engineering},
volume = {19},
number = {5},
abstract = {{Objective. Accurate decoding of surface electromyography (sEMG) is pivotal for muscle-to-machine-interfaces and their application e.g. rehabilitation therapy. sEMG signals have high inter-subject variability, due to various factors, including skin thickness, body fat percentage, and electrode placement. Deep learning algorithms require long training time and tend to overfit if only few samples are available. In this study, we aim to investigate methods to calibrate deep learning models to a new user when only a limited amount of training data is available. Approach. Two methods are commonly used in the literature, subject-specific modeling and transfer learning. In this study, we investigate the effectiveness of transfer learning using weight initialization for recalibration of two different pretrained deep learning models on new subjects data and compare their performance to subject-specific models. We evaluate two models on three publicly available databases (non invasive adaptive prosthetics database 2–4) and compare the performance of both calibration schemes in terms of accuracy, required training data, and calibration time. Main results. On average over all settings, our transfer learning approach improves 5%-points on the pretrained models without fine-tuning, and 12%-points on the subject-specific models, while being trained for 22% fewer epochs on average. Our results indicate that transfer learning enables faster learning on fewer training samples than user-specific models. Significance. To the best of our knowledge, this is the first comparison of subject-specific modeling and transfer learning. These approaches are ubiquitously used in the field of sEMG decoding. But the lack of comparative studies until now made it difficult for scientists to assess appropriate calibration schemes. Our results guide engineers evaluating similar use cases.},
keywords = {BCI, Computational Complexity, Deep Transfer-Learning, Machine Learning, transfer learning},
pubstate = {published},
tppubtype = {article}
}
@inproceedings{xavierfidencioClosedloopAdaptationBrainmachine2022,
title = {Closed-Loop Adaptation of Brain-Machine Interfaces Using Error-Related Potentials and Reinforcement Learning},
author = {Aline Xavier Fidencio and Christian Klaes and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2022.136},
year = {2022},
date = {2022-09-15},
urldate = {2022-09-15},
booktitle = {BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{sziburisDataset3DHand2022,
title = {A Dataset of 3D Hand Transport Trajectories Determined by Inertial Measurements from a Single Sensor},
author = {Tim Sziburis and Susanne Blex and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2022.186},
year = {2022},
date = {2022-09-15},
urldate = {2022-09-15},
booktitle = {BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{schmidtLinkingMuscleActivity2022,
title = {Linking Muscle Activity and Motion Trajectory},
author = {Marie Dominique Schmidt and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2022.191},
year = {2022},
date = {2022-09-15},
urldate = {2022-09-15},
booktitle = {BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{lehmlerModelingSubjectSpecfic2022,
title = {Modeling Subject Specfic Surface EMG Features by Means of Deep Learning},
author = {Stephan Johann Lehmler and Muhammad Saif-ur-Rehman and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2022.309},
year = {2022},
date = {2022-09-15},
urldate = {2022-09-15},
booktitle = {BC22 : Computational Neuroscience & Neurotechnology Bernstein Conference 2022},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@article{xavierfidencioErrorrelated,
title = {Error-Related Potentials in Reinforcement Learning-Based Brain-Machine Interfaces},
author = {Aline Xavier Fidencio and Christian Klaes and Ioannis Iossifidis},
url = {https://www.frontiersin.org/article/10.3389/fnhum.2022.806517},
doi = {https://doi.org/10.3389/fnhum.2022.806517},
year = {2022},
date = {2022-06-24},
urldate = {2022-06-24},
journal = {Frontiers in Human Neuroscience},
volume = {16},
abstract = {The human brain has been an object of extensive investigation in different fields. While several studies have focused on understanding the neural correlates of error processing, advances in brain-machine interface systems using non-invasive techniques further enabled the use of the measured signals in different applications. The possibility of detecting these error-related potentials (ErrPs) under different experimental setups on a single-trial basis has further increased interest in their integration in closed-loop settings to improve system performance, for example, by performing error correction. Fewer works have, however, aimed at reducing future mistakes or learning. We present a review focused on the current literature using non-invasive systems that have combined the ErrPs information specifically in a reinforcement learning framework to go beyond error correction and have used these signals for learning.},
keywords = {BCI, EEG, error-related potentials, Machine Learning, Reinforcement learning},
pubstate = {published},
tppubtype = {article}
}
@article{aliConTraNetSingleEndtoend2022,
title = {ConTraNet: A Single End-to-End Hybrid Network for EEG-based and EMG-based Human Machine Interfaces},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {http://arxiv.org/abs/2206.10677},
doi = {10.48550/arXiv.2206.10677},
year = {2022},
date = {2022-06-21},
urldate = {2022-06-21},
abstract = {Objective: Electroencephalography (EEG) and electromyography (EMG) are two non-invasive bio-signals, which are widely used in human machine interface (HMI) technologies (EEG-HMI and EMG-HMI paradigm) for the rehabilitation of physically disabled people. Successful decoding of EEG and EMG signals into respective control command is a pivotal step in the rehabilitation process. Recently, several Convolutional neural networks (CNNs) based architectures are proposed that directly map the raw time-series signal into decision space and the process of meaningful features extraction and classification are performed simultaneously. However, these networks are tailored to the learn the expected characteristics of the given bio-signal and are limited to single paradigm. In this work, we addressed the question that can we build a single architecture which is able to learn distinct features from different HMI paradigms and still successfully classify them. Approach: In this work, we introduce a single hybrid model called ConTraNet, which is based on CNN and Transformer architectures that is equally useful for EEG-HMI and EMG-HMI paradigms. ConTraNet uses CNN block to introduce inductive bias in the model and learn local dependencies, whereas the Transformer block uses the self-attention mechanism to learn the long-range dependencies in the signal, which are crucial for the classification of EEG and EMG signals. Main results: We evaluated and compared the ConTraNet with state-of-the-art methods on three publicly available datasets which belong to EEG-HMI and EMG-HMI paradigms. ConTraNet outperformed its counterparts in all the different category tasks (2-class, 3-class, 4-class, and 10-class decoding tasks). Significance: The results suggest that ConTraNet is robust to learn distinct features from different HMI paradigms and generalizes well as compared to the current state of the art algorithms.},
keywords = {BCI, Machine Learning, neural processing, signal processing},
pubstate = {published},
tppubtype = {article}
}
@inproceedings{doliwaDevelopmentScalableAnalogaccepted,
title = {Development of a Scalable Analog Front-End for Brain-Computer Interfaces},
author = {Sebastian Doliwa and Andreas Erbeslöh and Karsten Seidl and Ioannis Iossifidis},
year = {2022},
date = {2022-06-15},
urldate = {2022-06-15},
booktitle = {17th International Conference on PhD Research in Microelectronics and Electronics},
publisher = {IEEE Prime 2022},
address = {Sardinia, Italy},
keywords = {BCI, Implantable BCI},
pubstate = {published},
tppubtype = {inproceedings}
}
@article{aliAnchoredSTFTGNAAExtension2021a,
title = {Enhancing the decoding accuracy of EEG signals by the introduction of anchored-STFT and adversarial data augmentation method},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Susanne Dyck and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
url = {https://www.nature.com/articles/s41598-022-07992-w},
doi = {https://doi.org/10.1038/s41598-022-07992-w},
issn = {2045-2322},
year = {2022},
date = {2022-03-10},
urldate = {2022-03-10},
journal = {Nature Scientific Reports},
volume = {12},
issue = {1},
pages = {4245},
abstract = {Brain-computer interfaces (BCIs) enable communication between humans and machines by translating brain activity into control commands. Electroencephalography (EEG) signals are one of the most used brain signals in non-invasive BCI applications but are often contaminated with noise. Therefore, it is possible that meaningful patterns for classifying EEG signals are deeply hidden. State-of-the-art deep-learning algorithms are successful in learning hidden, meaningful patterns. However, the quality and the quantity of the presented inputs is pivotal. Here, we propose a novel feature extraction method called anchored Short Time Fourier Transform (anchored-STFT), which is an advanced version of STFT, as it minimizes the trade-off between temporal and spectral resolution presented by STFT. In addition, we propose a novel augmentation method, called gradient norm adversarial augmentation (GNAA). GNAA is not only an augmentation method but is also used to harness adversarial inputs in EEG data, which not only improves the classification accuracy but also enhances the robustness of the classifier. In addition, we also propose a new CNN architecture, namely Skip-Net, for the classification of EEG signals. The proposed pipeline outperforms all state-of-the-art methods and yields an average classification accuracy of 90.7 % and 89.54 % on BCI competition II dataset III and BCI competition IV dataset 2b, respectively.},
keywords = {Adversarial NN, BCI, computer science, EEG, Machine Learning, Quantitative Biology, Quantitative Methods},
pubstate = {published},
tppubtype = {article}
}
@article{schmidt2022motion,
title = {From Motion to Muscle},
author = {Marie Dominique Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
doi = {https://doi.org/10.48550/arXiv.2201.11501},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
journal = {arXiv: 2201.11501 [cs.LG]},
keywords = {BCI, Machine Learning, movement model, muscle signal generator, recurrent neural network},
pubstate = {published},
tppubtype = {article}
}
@inproceedings{fidencioErrorrelatedPotentialsDetection2022,
title = {Error-Related Potentials Detection with Dry- and Wet-Electrode EEG},
author = {Aline Xavier Fidencio and Tobias Glasmachers and Ioannis Iossifidis},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
booktitle = {FENS, Forum 2022},
publisher = {FENS, Federation of European Neuroscience Societies},
abstract = {Electroencephalography (EEG) is a non-invasive technique for measuring brain electrical activity from electrodes placed on the scalp surface. Improvements in this technology are particularly relevant because they also boost brain-machine interfaces (BMI) development. Commonly, gel-based electrodes are used since they guarantee a high-quality signal. Alternatively, dry electrodes have been introduced, more suitable for daily use. In this work, we compare conventional dry and wet electrode systems specifically for the detection of error-related potentials (ErrPs). ErrPs are elicited as a reaction to both self-made and external errors. There has been increased interest in the integration of these signals into BMIs to improve their performance since they provide a convenient source of feedback to the system with no extra workload for the subject. These signals can be used, e.g., to correct errors or even for system adaptation. ErrP-based BMIs in the literature have consistently used wet electrodes. Therefore, even though both electrodes types have been compared for other event-related potentials (e.g., P300), it is relevant to know whether the signal quality for the detection of ErrPs is comparable among them. In this work, we implement a simple game to elicit ErrPs and compare the quality of the measured signals. We tested the feasibility of the experimental protocol to elicit ErrP and the measured ErrP displayed a similar waveshape in terms of observed peaks. However, differences exist in both latencies as well as in their amplitude. These variations and other relevant characteristics have to be further verified with more subjects},
keywords = {BCI, EEG, error-related potentials, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{schmidtMotionIntentionPrediction2022a,
title = {Motion Intention Prediction},
author = {Marie Dominique Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
booktitle = {FENS, Forum 2022},
publisher = {FENS, Federation of European Neuroscience Societies},
abstract = {Motion intention prediction is the key to robot-assisted rehabilitation systems. These can rely on various biological signals. One commonly used signal is the muscle activity measured by an electromyogram that occurs between 50-100 milliseconds before the actual movement, allowing a real-world application to assist in time. We show that upper limb motion can be estimated from the corresponding muscle activity. To this end, eight-arm muscles are mapped to the joint angle, velocity, and acceleration of the shoulder, elbow, and wrist. For this purpose, we specifically develop an artificial neural network that estimates complex motions involving multiple upper limb joints. The network model is evaluated concerning its ability to generalize across subjects as well as for new motions. This is achieved through training on multiple subjects and additional transfer learning methods so that the prediction for new subjects is significantly improved. In particular, this is beneficial for a robust real-world application. Furthermore, we investigate the importance of the different parameters such as angle, velocity, and acceleration for simple and complex motions. Predictions for simple motions along with the main components of complex motions achieve excellent accuracy while joints that do not play a dominant role during the motion have comparatively lower accuracy.},
keywords = {BCI, Machine Learning, movement model, muscle signal generator, recurrent neural network},
pubstate = {published},
tppubtype = {inproceedings}
}
2021
@inproceedings{lehmlerTransferLearningPatientSpecific2021b,
title = {Transfer-Learning for Patient Specific Model Re-Calibration: Application to sEMG-Classification},
author = {Stephan Johann Lehmler and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2021.p005},
year = {2021},
date = {2021-10-01},
urldate = {2021-10-01},
publisher = {Bernstein Conferen},
keywords = {BCI, Computational Complexity, Deep Transfer-Learning, Machine Learning, transfer learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{schmidtArtificiallyGeneratedMuscle2021b,
title = {Artificially Generated Muscle Signals},
author = {Marie Dominique Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2021.p111},
year = {2021},
date = {2021-10-01},
urldate = {2021-10-01},
publisher = {Bernstein Conference},
keywords = {BCI, Machine Learning, movement model, muscle signal generator, recurrent neural network},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{xavierfidencioErrorCorrectionIntegration2021b,
title = {Beyond Error Correction: Integration of Error-Related Potentials into Brain-Computer Interfaces for Improved Performance},
author = {Aline Xavier Fidencio and Tobias Glasmachers and Christian Klaes and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2021.p163},
year = {2021},
date = {2021-10-01},
urldate = {2021-10-01},
publisher = {Bernstein Conference},
keywords = {BCI, error-related potentials, Machine Learning, Reinforcement learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{schmidtArtificiallyGeneratedMuscle2021,
title = {Artificially Generated Muscle Signals},
author = {Marie Dominique Schmidt and Tobias Glasmachers and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2021.p111},
year = {2021},
date = {2021-09-15},
urldate = {2021-09-15},
booktitle = {BC21 : Computational Neuroscience & Neurotechnology Bernstein Conference 2021},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{lehmlerTransferLearningPatientSpecific2021,
title = {Transfer-Learning for Patient Specific Model Re-Calibration: Application to sEMG-Classification},
author = {Stephan Johann Lehmler and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2021.p005},
year = {2021},
date = {2021-09-15},
urldate = {2021-09-15},
booktitle = {BC21 : Computational Neuroscience & Neurotechnology Bernstein Conference 2021},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning},
pubstate = {published},
tppubtype = {inproceedings}
}
@inproceedings{sziburisModellingGenerationHuman2021,
title = {Modelling the Generation of Human Upper-Limb Reaching Trajectories: An Extended Behavioural Attractor Dynamics Approach},
author = {Tim Sziburis and Susanne Blex and Tobias Glasmachers and Inaki Rano and Ioannis Iossifidis},
doi = {10.12751/nncn.bc2021.p078},
year = {2021},
date = {2021-09-15},
urldate = {2021-09-15},
booktitle = {BC21 : Computational Neuroscience & Neurotechnology Bernstein Conference 2021},
publisher = {BCCN Bernstein Network Computational Network},
keywords = {BCI, Machine Learning, movement model},
pubstate = {published},
tppubtype = {inproceedings}
}
@article{aliAnchoredSTFTGNAAExtension2021,
title = {Anchored-STFT and GNAA: An Extension of STFT in Conjunction with an Adversarial Data Augmentation Technique for the Decoding of Neural Signals},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Susanne Dyck and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
year = {2021},
date = {2021-08-01},
urldate = {2021-08-01},
journal = {arXiv:2011.14694 [cs, q-bio]},
abstract = {Brain-computer interfaces (BCIs) enable communication between humans and machines by translating brain activity into control commands. Electroencephalography (EEG) signals are one of the most used brain signals in non-invasive BCI applications but are often contaminated with noise. Therefore, it is possible that meaningful patterns for classifying EEG signals are deeply hidden. State-of-the-art deep-learning algorithms are successful in learning hidden, meaningful patterns. However, the quality and the quantity of the presented inputs is pivotal. Here, we propose a novel feature extraction method called anchored Short Time Fourier Transform (anchored-STFT), which is an advanced version of STFT, as it minimizes the trade-off between temporal and spectral resolution presented by STFT. In addition, we propose a novel augmentation method, called gradient norm adversarial augmentation (GNAA). GNAA is not only an augmentation method but is also used to harness adversarial inputs in EEG data, which not only improves the classification accuracy but also enhances the robustness of the classifier. In addition, we also propose a new CNN architecture, namely Skip-Net, for the classification of EEG signals. The proposed pipeline outperforms all state-of-the-art methods and yields an average classification accuracy of 90.7 % and 89.54 % on BCI competition II dataset III and BCI competition IV dataset 2b, respectively.},
keywords = {BCI, Machine Learning, Quantitative Biology, Quantitative Methods},
pubstate = {published},
tppubtype = {article}
}
@article{lehmler2021deep,
title = {Deep Transfer-Learning for patient specific model re-calibration: Application to sEMG-Classification},
author = {Stephan Johann Lehmler and Muhammad Saif-ur-Rehman and Tobias Glasmachers and Ioannis Iossifidis},
url = {https://api.semanticscholar.org/CorpusID:245634948},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
keywords = {BCI, Computational Complexity, Deep Transfer-Learning, Machine Learning},
pubstate = {published},
tppubtype = {article}
}
2020
@article{10.1088/1741-2552/abc8d4,
title = {SpikeDeep-Classifier: A deep-learning based fully automatic offline spike sorting algorithm},
author = {Muhammad Saif-ur-Rehman and Omair Ali and Susanne Dyck and Robin Lienkämper and Marita Metzler and Yaroslav Parpaley and Jörg Wellmer and Charles Liu and Brian Lee and Spencer Kellis and Richard Andersen and Ioannis Iossifidis and Tobias Glasmachers and Christian Klaes},
url = {http://iopscience.iop.org/article/10.1088/1741-2552/abc8d4},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Journal of Neural Engineering},
abstract = {Objective. Advancements in electrode design have resulted in micro-electrode arrays with hundreds of channels for single cell recordings. In the resulting electrophysiological recordings, each implanted electrode can record spike activity (SA) of one or more neurons along with background activity (BA). The aim of this study is to isolate SA of each neural source. This process is called spike sorting or spike classification. Advanced spike sorting algorithms are time consuming because of the human intervention at various stages of the pipeline. Current approaches lack generalization because the values of hyperparameters are not fixed, even for multiple recording sessions of the same subject. In this study, a fully automatic spike sorting algorithm called “SpikeDeep-Classifier” is proposed. The values of hyperparameters remain fixed for all the evaluation data. Approach. The proposed approach is based on our previous study (SpikeDeeptector) and a novel background activity rejector (BAR), which are both supervised learning algorithms and an unsupervised learning algorithm (K-means). SpikeDeeptector and BAR are used to extract meaningful channels and remove BA from the extracted meaningful channels, respectively. The process of clustering becomes straight-forward once the BA is completely removed from the data. Then, K-means with a predefined maximum number of clusters is applied on the remaining data originating from neural sources only. Lastly, a similarity-based criterion and a threshold are used to keep distinct clusters and merge similar looking clusters. The proposed approach is called cluster accept or merge (CAOM) and it has only two hyperparameters (maximum number of clusters and similarity threshold) which are kept fixed for all the evaluation data after tuning. Main Results. We compared the results of our algorithm with ground-truth labels. The algorithm is evaluated on data of human patients and publicly available labeled non-human primates (NHPs) datasets. The average accuracy of BAR on datasets of human patients is 92.3% which is further reduced to 88.03% after (K-means + CAOM). In addition, the average accuracy of BAR on a publicly available labeled dataset of NHPs is 95.40% which reduces to 86.95% after (K-mean + CAOM). Lastly, we compared the performance of the SpikeDeep-Classifier with two human experts, where SpikeDeep-Classifier has produced comparable results. Significance. The results demonstrate that “SpikeDeep-Classifier” possesses the ability to generalize well on a versatile dataset and henceforth provides a generalized well on a versatile dataset and henceforth provides a generalized and fully automated solution to offline spike sorting.},
keywords = {BCI, CNN, Machine Learning, Spike Sorting},
pubstate = {published},
tppubtype = {article}
}
@article{ali2020improving,
title = {Improving the performance of EEG decoding using anchored-STFT in conjunction with gradient norm adversarial augmentation},
author = {Omair Ali and Muhammad Saif-ur-Rehman and Susanne Dyck and Tobias Glasmachers and Ioannis Iossifidis and Christian Klaes},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {arXiv preprint arXiv:2011.14694},
keywords = {Adversarial NN, BCI, EEG, Machine Learning},
pubstate = {published},
tppubtype = {article}
}
2019
@misc{ur-reimann2019a,
title = {Universal SpikeDeeptector},
author = {Muhammad Saif-ur-Rehman and Robin Lienkämper and Susanne Dyck and A Rayana and Y Parpaley and J Wllner and Charles Liu and Brian Lee and Spencer Kellis and D Manahan-Vaughn and O Güntürkün and Richard Andersen and Ioannis Iossifidis and Tobias Glasmachers and Christian Klaes},
year = {2019},
date = {2019-01-01},
urldate = {2019-01-01},
publisher = {SfN 2019},
abstract = {State-of-the-art microelectrode array technology enables simultaneous, large-scale single unit recordings from hundreds of channels. Identification of channels recording neural data as compared to noise is the first step for all further analyses. Automatizing this process aims at minimizing the human involvement and time for manual curation. In our previous study, we introduced the “SpikeDeeptector” (SD), which enables us to automatically detect and track channels containing neural data from different human patients implanted with different types of microelectrodes across different brain areas. SD works on human data and to some extent on the data of non-human primates (NHPs). However, to make SD more versatile we proposed a more generalized method called “Universal SpikeDeeptector (USD)”, which is an extended version of SD. USD intends to detect and track the channels containing neural data recorded from four different species (rats, ravens, NHPs and humans) using different kinds of microelectrodes and different recording sites. To our knowledge, there is no method that can simultaneously detect and track neural data of multiple species. To enable contextual learning, USD constructs a feature vector from a batch of waveforms. The constructed feature vectors are then fed into a deep-learning algorithm, which learns contextualized, temporal and spatial patterns. USD is a supervised learning method. Therefore, it requires labeled data for training. It is mainly trained on data from a single human tetraplegic patient, and a small but equal portion of data from the remaining three species. The trained model is then evaluated on a test dataset collected from several humans, NHPs, rats, and birds. The results show that the USD performed consistently well across data collected from each species.},
keywords = {BCI, CNN, Machine Learning, Spike Detection, Spike Sorting},
pubstate = {published},
tppubtype = {misc}
}
@article{Saif-ur-Rehman2019,
title = {SpikeDeeptector: a deep-learning based method for detection of neural spiking activity},
author = {Muhammad Saif-ur-Rehman and Robin Lienkämper and Yaroslav Parpaley and Jörg Wellmer and Charles Liu and Brian Lee and Spencer Kellis and Richard Andersen and Ioannis Iossifidis and Tobias Glasmachers and Christian Klaes},
url = {https://iopscience.iop.org/article/10.1088/1741-2552/ab1e63/meta},
doi = {10.1088/1741-2552/ab1e63},
year = {2019},
date = {2019-01-01},
urldate = {2019-01-01},
journal = {Journal of Neural Engineering},
volume = {16},
number = {5},
pages = {056003},
abstract = {Objective . In electrophysiology, microelectrodes are the primary source for recording neural data (single unit activity). These microelectrodes can be implanted individually or in the form of arrays containing dozens to hundreds of channels. Recordings of some channels contain neural activity, which are often contaminated with noise. Another fraction of channels does not record any neural data, but only noise. By noise, we mean physiological activities unrelated to spiking, including technical artifacts and neural activities of neurons that are too far away from the electrode to be usefully processed. For further analysis, an automatic identification and continuous tracking of channels containing neural data is of great significance for many applications, e.g. automated selection of neural channels during online and offline spike sorting. Automated spike detection and sorting is also critical for online decoding in brain–computer interface (BCI) applications, in which on...},
keywords = {BCI, CNN, Data Reduction, Machine Learning, Spike Sorting},
pubstate = {published},
tppubtype = {article}
}
2017
@misc{Iossifidis2017b,
title = {Low dimensional representation of human arm movement for efficient neuroprosthetic control by individuals with tetraplegia},
author = {Ioannis Iossifidis and Christian Klaes},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
publisher = {SfN 2017},
abstract = {Over the last decades the generation mechanism and the representation of goal- directed movements has been a topic of intensive neurophysiological research. The investigation in the motor, premotor, and parietal areas led to the discovery that the direction of hand's movement in space was encoded by populations of neurons in these areas together with many other movement parameters. These distributions of population activation reflect how movements are prepared ahead of movement initiation, as revealed by activity induced by cues that precede the imperative signal (Georgopoulos, 1991). Inspired by those findings a model based on dynamical systems was proposed both, to model goal directed trajectories in humans and to generate trajectories for redundant anthropomorphic robotic arms. The analysis of the attractor dynamics based on the qualitative comparison with measurements of resulting trajectories taken from arm movement experiments with humans (Grimme u. a., 2012) created a framework able to reproduce and to generate naturalistic human like arm trajectories (Iossifidis und Rano, 2013; Iossifidis, Schöner u. a., 2006). The main idea of the methodology is to choose low-dimensional, behavioral va- riables of the goal task can be represented as attractor states of those variables. The movement is generated through a dynamical system with attractors and repellers on the behavioral space, at the goal and constraint positions respectively. When the motion of the robot evolves according to the dynamics of these systems, the behavioral variables will be stabilized at their attractors. Movement is represented by the polar coordinates $phi$,$theta$ of the movement direction (heading direction) and the angular frequency $ømega$ of a hopf oscillator, generating the velocity profile of the arm movement. Therefore, the system dynamics will be expressed in terms of these variables. The target and each obstacle induce vector fields over these variables in a way that states where the hand is moving closer to the target are attractive, while states where it is moving towards an obstacle are repellant. Contributions from different sources are weighted by different factors, e.g. in the vicinity of an obstacle, the contribution from that obstacle must dominate the behavior to guarantee constraint satisfaction (collision prevention). Based on three parameters the presented framework is able to generate temporal stabilized (timed) discrete movements, dealing with disturbances and maintaining an approximately constant movement time. In the current study we will implant two 96-channel intracortical microelectrode arrays in the primary motor and the posterior parietal cortex (PPC) of an individual with tetraplegia. In the training phase the parameters of the dynamical systems will be tuned and optimized by machine learning algorithms. Rather controlling directly the arm movement and adjusting continuously parameters, the patient adjust by his or hers thoughts the three parameters of the dynamics, which remain almost constant during the movement. Only when the motion plan is changing the parameters have to be readjusted. The target directed trajectory evolves from the attractor solution of the dynamical systems equations, which means that the trajectory is generated while the system is in a stable stationary state, a fixed-point attractor. The increase of the degree of assistance lowers the cognitive load of the patient and enables the acknowledgement of the desired task without frustration. In addition we aim to replace the robotic manipulator by an exoskeleton for the upper body which will enable the patients to move his or hers own limbs, which would complete the development of a real neuroprosthetic device for every day use.},
keywords = {BCI, Dynamical systems, movement model, neuroprosthetic, Robotics},
pubstate = {published},
tppubtype = {misc}
}
@conference{nokey,
title = {Low dimensional representation of human arm movement for efficient neuroprosthetic control by individuals with tetraplegia},
author = {Christian Klaes and Ioannis Iossifidis},
year = {2017},
date = {2017-01-01},
urldate = {2017-01-01},
booktitle = {SfN Meeting 2017},
keywords = {BCI, Dynamical systems, movement model, neuroprosthetic, Robotics},
pubstate = {published},
tppubtype = {conference}
}